SnapFast™ Restriction Site Functions
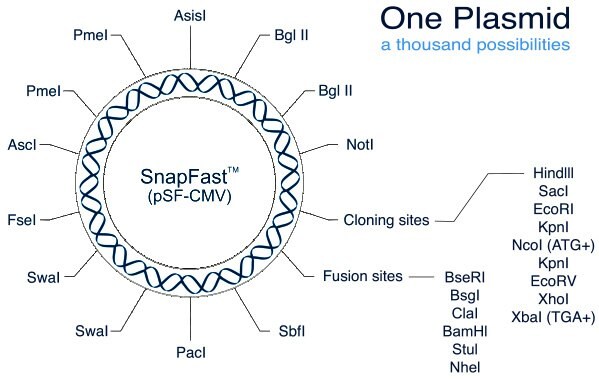
All DNA sections are pre-screened, and where possible modified, to remove any of the restriction sites found within the core SnapFast plasmids to maintain their flexibility. If a DNA section still contains a site this will be stated on the individual product data sheet.
AsiSI / SgfI - GCGAT'CGC
The AsiSI / SbfI site has two primary functions. SgfI is an isoschizomer of AsiSI.
Function 1: Join Multiple Expression Cassettes Togther / Concatemerise Genes
If you would like to insert two gene cassettes together into the same SnapFastTM plasmid this can be achieved using the AsiSI restriction site in conjuntion with the PacIsite. In order to do this, one of your SnapFast vectors must be linearised with either AsiSI or PacI, and the second vector must be cut with both AsiSI and PacI. The latter excision will remove your entire gene cassette from the second vector. This fragment can then be ligated into the first vector that was linearised with either AsiSI or PacI. This is possible because both AsiSI and PacI have compatible cohesive ends.
This cloning strategy will ablate one of the ligated sites by creating an AsiSI/PacI hybrid site that can no longer be cut by either enzyme. If you screen to ensure that both of your genes are in the same orientation as when they started, it should be possible to repeat the process with even more genes, because the ablation process will result in the reconstitution of a single AsiSI and PacI site in the vector (figure 1). This will enable you to repeat the whole process again with more genes.
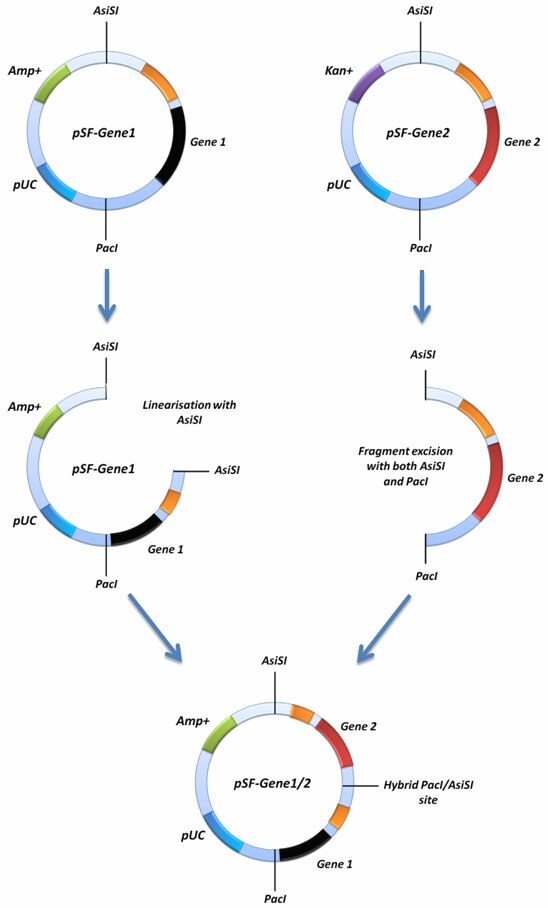
Figure 1. Diagramatical representation of the concatemerisation function.
For this function to work, you will need to confirm that you have no PacI or AsiSI sites present in your plasmids, other than those shown on our maps. Some genes you may have inserted may contain these sites. If one of your plasmids contains an additional site, it is still possible to perform the process because one of the plasmids only requires linearisation with one enzyme.
Function 2: Viral genomic insertion
We are designing a range of viral vectors containing AsiSI and PacI/NotI/NheI/SbfI sites. This will allow the direct cloning of your expression cassettes into each of these viruses. The AsiSI and PacI sites are designed to be used to insert a gene (or concatemerised genes), including the promoters, into the viral vectors.
Bgl2 - A'GATCT
Primary Function: Promoter Insertion
The Bgl2 site always flanks our promoters or promoter multiple cloning sites. We recommend using the closest unique restriction sites (e.g. AsiSI and NotI) before attempting to use the Bgl2 sites. This will increase the chances of your ligation yielding clones in the correct orientation. If you would like to insert your own promoter then you can use our promoter specific multiple cloning site plasmids (PromMCS).
NotI - GC'GGCCGC
The NotI site has three functions.
Function 1: Cap Independent translation
Using the NotI site in conjunction with the downstream NcoI site, it is possible to allow cap independent translation of your gene using one of our IRES inserts.
Function 2: Insertion of 5' fusion sequences and peptide tags
The NotI site can be used to allow the insertion of coding sequences that are designed to be fused with your gene of interest. For example:
Secretory signal peptide tags or purification/affinity peptide tags.
Function 3: Viral genomic insertion
We are currently designing a range of viral vectors containing a NotI site upstream of SbfI and PacI sites. These sites allow the direct cloning of your SnapFast genes into these viral vectors. The NotI site is designed to be used for inserting genes into viruses where a promoter is not required (e.g. RNA viruses or viruses already containing a promoter).
HindIII - A'AGCTT
The HindIII site has two primary functions.
Function 1: Insertion of 5' fusion sequences
The HindIII site can be used to allow the insertion of coding sequences that are designed to be fused with your gene of interest. For example:
Secretory signal peptide tags or purification/affinity peptide tags.
Function 2: Gene Cloning
As the 5' gene cloning site:
HindIII is part of the multiple cloning site (polylinker) and can be used to insert genes. The built in Kozak and Shine-Dalgarno ribosomal entry sequences within the SnapFast multiple cloning site are downstream of HindIII. It will therefore need to be added by your gene insertion to enable efficient translation.
The HindIII restriction site is also found in other commonly used vectors including TOPO and pGL3 based vectors. This allows cloning of your genes directly into our plasmids.
As the 3' gene cloning site:
The HindIII site can be used as the 3' cloning site for your gene. We recommend using the XbaI (stop codon TAG+) site as the 3' cloning site where possible, as this will allow the greatest flexibility in future SnapFast cloning.
SacI - GAGCT'C
The SacI site has one function.
Function: Gene Cloning
As the 5' gene cloning site:
SacI is part of the multiple cloning site (polylinker) and can be used to insert genes, although we normally recommend using the NcoI (ATG+) site as your 5' cloning site where possible. This will provide you with the greatest flexibility for future SnapFast cloning. However, the limitations of using SacI generally refer only to N-terminal protein tags that are aligned with the NcoI restriction sites, but we also sell these same tags in the NotI/HindIII position as well, and most peptide tags should still be compatible.
The SacI restriction site is also found in commonly used vectors, including T0P0 based vectors. This allows cloning of your gene directly from those vectors into the SnapFast system.
As the 3' gene cloning site:
The SacI site can be used as the 3' cloning site, however, a KOZAK sequence or Shine-Dalgarno sequence should be placed at the 5' end of your gene, depending your expression requirements. We recommend using the XbaI (stop codon TAG+) site as the 3' cloning site where possible, to allow you the greatest flexibility in future SnapFast cloning.
EcoRI - G'AATTC
The EcoRI site has one function.
Function: Gene Cloning
As the 5' gene cloning site:
EcoRI is part of the multiple cloning site (polylinker) and is designed to be used to insert genes into the SnapFast vector. The built in KOZAK and Shine-Dalgarno sequences within the SnapFast system are downstream of this site, and should therefore be restored by your gene insertion to enable efficient translation in your intended expression system. We recommend using the NcoI (ATG+) site as your 5' cloning site where possible. This will provide you with the greatest flexibility for future SnapFast cloning. The limitations of using EcoRI generally refer only to N-terminal protein tags that are NcoI aligned, but we also sell these same tags in the NotI/HindIII position as well, and most peptide tags should still be compatible.
The EcoRI restriction site is also found in commonly used vectors including T0P0 based vectors. This allows cloning directly from those vectors into the SnapFast system.
As the 3' gene cloning site:
The EcoRI site can be used as the 3' cloning site, however, a KOZAK sequence or Shine-Dalgarno sequence should be placed at the 5' end of your gene depending your expression system. We recommend using the XbaI (stop codon TAG+) site as the 3' cloning site where possible to allow you the greatest flexibility in future SnapFast cloning.
If your gene contains an EcoRI site, try amplifiying to add a MfeI site. These sites produce compatible cohesive (sticky) ends.
KpnI - GGTAC'C
The KpnI sites (there are two in the vector) have two functions.
Function 1: Removal of the ATG start codon
There are two KpnI sites in the SnapFast MCS. These sites allow the removal of the start codon within the NcoI site that they flank. This then allows downstream sites (e.g. EcoRV or XhoI) to be used instead to insert a gene, without the concern of premature ribosomal translation initiation at the start codon in the NcoI site.
To remove the NcoI site simply cut the SnapFast vector with KpnI and re-circularise it without de-phosphorylation. Diagnostic restriction of recombinants with NcoI should confirm if the site is missing, demonstrated by a lack of linearisation. Genes can then be cloned into the downstream restriction sites (EcoRV, XhoI) of the NcoI deleted SnapFast recombinants without premature translation initiation at the incorrect ATG codon. We also sell an NcoI deleted pSF-CMV vector.
Function 2: Gene Cloning
As the 5' gene cloning site:
KpnI is part of the multiple cloning site (polylinker) and is designed to be used to insert genes into the SnapFast vector. The built in KOZAK sequence within the SnapFast system is downstream of this site and should be restored by your gene insertion to enable efficient translation. The Shine-Dalgarno site is adjacent (upstream) to the KpnI site, and therefore your gene should ideally be positioned the correct number of base pairs downstream of this site if you intend to express it in bacteria. The last base of the Shine-Dalgarno sequence (AGGAGG) should be about 8 base pairs upstream from the first base of the start codon (ATG), although 4-7 can often also work fine.
We recommend using the NcoI (ATG+) site as your 5' cloning site where possible. This will provide you with the greatest flexibility for future SnapFast cloning, although most of our inserts should still be compatible when using the KpnI site as the 5' cloning site.
The KpnI restriction site is also commonly found in other expression vectors, including T0P0 based vectors. This allows cloning directly from those vectors into the SnapFast system.
As the 3' gene cloning site:
The KpnI site can be used as the 3' cloning site, but not in conjunction with NcoI, because NcoI is flanked by two KpnI sites. When using the KpnI site as the 3' cloning site it will alter the position of the KOZAK and Shine-Dalgarno (AGGAGG) sequences at the 5' end of your gene. If you are cloning your gene into KpnI alone please read the previous section on using the KpnI site as the 5' cloning site. We recommend using the XbaI (stop codon TAG+) site as the 3' cloning site where possible to allow you the greatest flexibility in future SnapFast cloning.
NcoI (Start - ATG+) - C'CATGG
The NcoI site has one function.
Function: Gene cloning with a built-in KOZAK consensus sequence and start codon
The NcoI site contains the pre-designed start codon for the SnapFast system, and contains a consensus KOZAK sequence. It is therefore the easiest point at which to insert your gene, leaving you the most options and flexibility for subsequent SnapFast modifications. There is also a consensus Shine-Dalgarno sequence positioned the correct number of base pairs upstream of the start codon to enable efficient expression in bacteria.
If you intend to PCR amplify your gene before inserting it into the NcoI site, you should ensure that the start codon of your gene is within the NcoI site. Placing the NcoI site upstream of your ATG start codon could result in a frame shift, or the presence of exogenous amino acids at the amino terminus of your protein product. We also sell an NcoI deleted pSF-CMV vector.
If you are using the NcoI site as the 5' cloning site we recommend using the XbaI site as the 3' cloning site and either placing your stop codon within the XbaI site (TAG+), or allowing your coding sequence to read through into one of the built-in stop codons in pSF-CMV-Amp. This provides you with the greatest flexibility for future SnapFast cloning.
The NcoI restriction site is also found in many other plasmid vectors. This allows cloning directly from those vectors into the SnapFast system.
If your gene contains a NcoI site, try PCR amplifiying it to add a BspHI, FatI or PciI site. These sites produce NcoI compatible cohesive (sticky) ends.
EcoRV - GAT'ATC
The EcoRV site has one function.
Function: Blunt Gene Cloning
As the 5' gene cloning site
EcoRV is part of the multiple cloning site (polylinker) and is designed to be used to insert genes into the SnapFast vector. In order to clone the 5' end of your gene into this position the SnapFast vector must be cleaved with KpnI, and re-circularised, to remove the start codon within the NcoI site. We also sell an NcoI deleted pSF-CMV vector. This will remove the built in KOZAK sequence which must be restored by your gene insertion to enable efficient translation. If you plan to express your gene in bacteria, the distance from the built-in Shine-Dalgarno sequence to your start codon must also be considered, please see the KpnI 'Function 2: gene cloning' section for more details.
As the 3' gene cloning site
The EcoRV site can be used as the 3' cloning site in conjunction with an upstream site (e.g. NcoI), however, we recommend using the XbaI (Stop Codon TAG+) site as the 3' cloning site where possible to allow you the greatest flexibility in future SnapFast cloning. If EcoRV is used alone, the guidlines for 5' EcoRV cloning must be followed. If you are not using NcoI as the upstream site you should ensure that your gene has a KOZAK or a Shine-Dalgarno sequence to allow ribosomal initiation in the expression system you intend to use.
The EcoRV restriction site is also found in commonly used vectors, including T0P0 based vectors. This allows cloning of your gene directly from those vectors into the SnapFast system. However, blunt cloning strategies have a higher failure rate than sticky ended cloning strategies, we recommend using sticky compatible cohesive end ligations where possible, but EcoRV can be used if necessary.
XhoI - C'TCGAG
The XhoI site has one function.
Function: Gene Cloning
As the 5' gene cloning site
We do not normally recommend using the XhoI site as a 5' cloning site because this makes the 5' UTR quite long, however, XhoI is part of the multiple cloning site (polylinker) and if necessary it can be used for this purpose. We have not validated its function in this position. In order to clone the 5' end of your gene into this position the SnapFast vector must be cleaved and re-circularised with KpnI to remove the start codon within the NcoI site. This will also remove the built in KOZAK sequence which should be restored by your gene insertion to enable efficient translation (in eukaryotic systems). If you plan to express your gene in bacteria, the distance from the built-in Shine-Dalgarno sequence to your start codon should also be considered, please see the KpnI 'Function 2: gene cloning' section for more details. We also sell an NcoI deleted pSF-CMV vector.
As the 3' gene cloning site
The XhoI site can be used as the 3' cloning site in conjunction with an upstream site (e.g. NcoI), however, we recommend using the XbaI (Stop Codon TAG+) site as the 3' cloning site where possible to allow you the greatest flexibility in future SnapFast cloning. If the 5' cloning site is EcoRV, or you are inserting into XhoI alone, then the start codon in the NcoI site must first be removed by cleaving and re-circularising with KpnI, although we also sell an NcoI deleted pSF-CMV vector that does not require this. Any 5' cloning site upstream of EcoRV can be used without removing the NcoI site when using XhoI as the 3' cloning site. Please also see the important note at the top of this section for information regarding the positioning of your genes stop codon.
The XhoI restriction site is also found in commonly used vectors, including T0P0 based vectors. This allows cloning directly from those vectors into the SnapFast system.
If your gene contains a XhoI site, try PCR amplifiying it to add a SalI site instead. These sites produce compatible cohesive (sticky) ends. The XhoI site in pSF-CMV-Amp is also recognised and cut by the eight base pair cutting enzyme AbsI (CC'TCGAGG) which produces the same sticky overhangs as XhoI.
XbaI (Stop - TAG+) - T'CTAGA
The XbaI site has one function.
Function: Gene cloning with a built-in stop codon
As the 5' gene cloning site:
We do not recommend using the XbaI site as a 5' cloning site because this makes the 5' UTR quite long, however, XbaI is part of the multiple cloning site (polylinker) and if necessary it can be used for this purpose. We have not validated its function in this position. In order to clone the 5' end of your gene into this position the SnapFast vector must be cleaved and re-circularised with KpnI to remove the start codon within the NcoI site, although we also sell an NcoI deleted pSF-CMV vector which does not require this. This will also remove the built in KOZAK sequence which should be restored by your gene insertion to enable efficient translation. The distance between the Shine-Dalgarno sequence and your start codon may also be incorrect if using the XbaI site as your 5' insertion site, please see the KpnI 'Function 2: gene cloning' section for more details.
As the 3' gene cloning site:
The XbaI site has been exploited to encode a built in stop codon (TGA), which is used by most of the genes we sell in our MCS. It is, therefore, the easiest point at which to insert your gene and leaves you the most options and flexibility for subsequent SnapFast modifications. If you intend to PCR amplify your gene before insertion into the XbaI site, it is optimal to include your start codon within the NcoI site if possible.
The use of the stop codon in the XbaI site allows the fusion of sequences to the C-terminus of your genes, such as peptide tags and reporter genes, using one of our Fuzyme sites (BseRI or BsgI, see below). These sites can be found in our fusion vectors, where the stop codon in the XbaI site has been ablated. They are non-palindromic restriction sites and cleave the DNA at a defined number of base pairs away from their binding sites.
If your gene contains a XbaI site, try PCR amplifiying it to add a SpeI, NheI or AvrII site to the end. These sites produce compatible cohesive (sticky) ends. Many of the sites in our vectors have similar options and flexibility, please see each site on this page for more information.
BseRI (Fuzyme site) - 'n(8)CTCCTC
BseRI has one function.
Function: Fusion of coding sequences to your gene
BseRI is a unique site in our vectors which has novel properties that add significant flexibility to our vectors. Within all SnapFast vectors, there are two sites that we call the Fuzyme sites (BseRI and BsgI). These sites allow you to fuse coding sequences downstream of your gene, provided you have positioned your stop codon as the TAG stop codon within the XbaI site. All of the genes we sell in the multiple cloning site will end with a TAG stop codon that can be cleaved by BseRI to enable the retrospective fusion of coding sequences.
By using our fuzyme system you can insert your gene with either no, or minimal, amino acids at the C-terminus, but then modify the gene by adding an additional coding sequence at a later date. This can be important for functional studies where the behaviour of the wild-type gene is preferred, but modified derivatives may subsequently be required.
How do Fuzyme sites work?:
BseRI is a member of the type IIS restriction group of enzymes that recognise a non-palindromic sequence, and then cleave the DNA at a site that is a defined number of nucleotides away from the original binding site, independent of the sequence at the cut position. In the case of BseRI, the enzyme cleaves 8 base pairs upstream from the first base of the BseRI site on the top strand in the sequence maps we provide, and 10 nucleotides upstream on the bottom strand. In almost all of our plasmids this results in a cleavage within the XbaI site between the first adenosine residue and the guanosine residue on the upper strand (positions 4 and 5 in the XbaI site) and between the equivalent positions in the palindromic XbaI site on the lower strand (between the first adenosine residue and the guanosine residue, 5' to 3' on the lower strand).
If your gene contains an XbaI site, or using an XbaI site alters the penultimate codon of your gene, try PCR amplifying it to add a SpeI, NheI or AvrII site to the end. These sites produce compatible cohesive (sticky) ends. The fuzyme system will still work after you have ligated in your product to the XbaI site, even though you have created a hybrid site, because BseRI will cut at the same position, regardless of the sequence in that region. The BseRI restriction enzyme is available from New England Biolabs.
BsgI (Fuzyme site) - 'n(13)CTGCAC
The BsgI site has one function.
Function: Fusion of coding sequences to your gene
BsgI is a unique site in our vectors which has novel properties that add significant flexibility to our vectors. Within all SnapFast vectors, there are two sites that we call the Fuzyme sites (BseRI and BsgI). These sites allow you to fuse coding sequences downstream of your gene, provided you have positioned your stop codon as the TAG stop codon within the XbaI site. These sites allow you to fuse coding sequences downstream of your gene, provided you have positioned your stop codon as the TAG stop codon within the XbaI site. All of the genes we sell in the multiple cloning site will end with a TAG stop codon that can be cleaved by BsgI to enable the retrospective fusion of coding sequences.
By using our fuzyme system you can insert your gene with either no, or minimal, amino acids at the C-terminus, but then modify the gene by adding an additional coding sequence at a later date. This can be important for functional studies where the behaviour of the wild-type gene is preferred, but modified derivatives may subsequently be required.
How do Fuzyme sites work:
BsgI is a member of the typeIIS restriction group of enzymes that recognise a non-palindromic sequence, and then cleave the DNA at a site that is a defined number of nucleotides away from the original binding site, independent of the sequence at the cut position. In the case of BsgI, the enzyme cleaves 14 base pairs upstream from the first base of the BsgI site on the top strand in the sequence maps we provide, and 16 nucleotides upstream on the bottom strand. In the parental vector this results in a cleavage within the XbaI site between the first adenosine residue and the guanosine residue on the upper strand (positions 4 and 5 in the XbaI site) and between the equivalent positions in the palindromic XbaI site on the lower strand (between the first adenosine residue and the guanosine residue, 5' to 3' on the lower strand.
If your gene contains an XbaI site, or using an XbaI site alters the penultimate codon of your gene, try PCR amplifying it to add a SpeI, NheI or AvrII site to the end. These sites produce compatible cohesive (sticky) ends. The fuzyme system will still work after you have ligated in your product to the XbaI site, even though you have created a hybrid site, because BseRI will cut at the same position, regardless of the sequence in that region. The BseRI restriction enzyme is available from New England Biolabs.
ClaI - AT'CGAT
The ClaI site has two functions.
Function 1: Insert an IRES or Second Promoter
The ClaI site can be used to insert either an IRES or second promoter to drive the expression of a second downstream gene. We recommend inserting the IRES into your vector using the ClaI site as the 5' site and the BamHI site as the 3 site, although any sites external to these sites will also work. IRESs can be selective about where they initiate translation from, for this reason we have positioned a PciI site in the correct position for this to occur. This site contains an ATG start codon that must be used. If a PciI site will not fit at the start of your gene, try to add an NcoI or BspHI site instead. When cleaved these enzyme sites produce the same overhang and can be ligated together.
Function 2: Adding a Peptide Tag
The ClaI site is always downstream of any C-terminal peptide tags that we sell in the main MCS. It can therefore be used insert peptide tags downstream of your gene of interest.
BamHI - G'GATCC
The BamHI site has one main function.
Function 1: Insert an IRES element or a Second Promoter
We have developed both IRES expression systems and second promoter expression systems that are driven from within the MCS. These are both normally flanked by BamHI as the 3' site to allow them to be ligated into the main MCS.
If you need to insert something into the BamHI site, but the sequence contains this site, try amplifiying to add a Bgl2 site. These sites produce compatible cohesive (sticky) ends and can be ligated together.
StuI - AGG'CCT
The StuI site simply functions a blunt site into which DNA sequences can be inserted. This can either be into the 3' UTR of the mRNA if only one gene is upstream, or it can be used to insert genes downstream of an IRES or second promoter using the relevant plasmid.
NheI - G'CTAGC
The NheI site has one primary function.
Function 1: Insert an IRES or second promoter gene cassette
We have developed a range of reporter genes driven by expresion from either an internal ribosome entry site (IRES) or second promoter. These cassettes always end with NheI as the 3' site.
SbfI - CCTGCA'GG
The SbfI site has two main functions.
Function 1: Insertion of an SV40 Origin of replication
The SbfI site is downstream of the SV40 poly A site (base pairs 1043 to 1234) and can be used to insert an SV40 origin of replication that allows plasmid maintanance in cells expressing the SV40 large T antigen.
Function 2: Insertion of an SV40 enhancer
The SbfI site can be used to insert an SV40 enhancer to increase the level of gene expression from a weak promoter. We have found that the addition of this sequence to promoters that are already strong (e.g. CMV) does not signficantly enhancer gene expression, however, it can improve weaker, less active promoters.
PacI - TTAAT'TAA
The PacI site has two main functions.
Function 1: Concatemerization
If you would like to insert two of your SnapFast gene cassettes into the same SnapFast vector this can be achieved using the AsiSI site in conjuntion with the PacIsite. In order to do this, one of your SnapFast vectors must be linearised with either AsiSI or PacI, and the other must be cut with both AsiSI and PacI. The latter should remove your gene expression cassette from the second vector and this fragment can be ligated into the first vector that was linearised with either AsiSI or PacI. This is possible because both AsiSI and PacI have compatible cohesive ends.
This cloning strategy, depending on the orientation of the insertion, ablates one of the ligated sites by creating an AsiSI/PacI hybrid site that can no longer be cut by either enzyme. If you screen to ensure that both genes are in the same orientation as when they started, it should be possible to repeat the process with even more genes because the ablation process will result in the reconstitution of a single AsiSI and PacI site in the vector. Allowing you to repeat the whole process again with more genes.
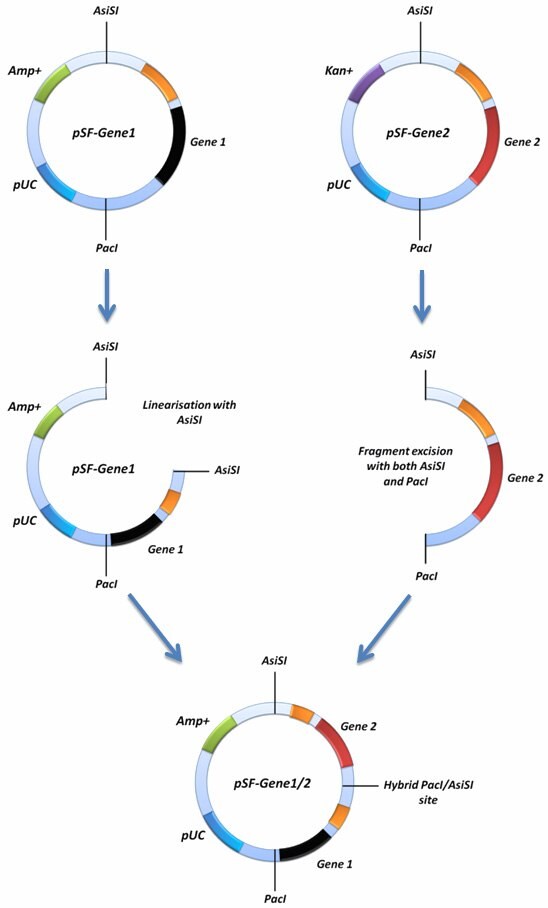
Figure 2. Diagramatical representation of the concatemerisation function of the SnapFast system. This method re-constitutes both the PacI and AsiSI sites, allowing multiple genes to be inserted into the same vector.
For this function to work, you will need to confirm that you have no PacI or AsiSI sites present in your vectors, other than those shown on our maps. Some genes you may have inserted may contain these sites. If one of your vectors contains an additional site, it is still possible to perform concatemerisation because one of the vectors only requires linearisation with only one enzyme. For example, if the gene in your starting plasmid contains an AsiSI site, you can linearise it with PacI, if your gene has a PacI site you can linearise it with AsiSI. The process is the same in each possibility, but just with the opposite enzyme, and concatemerising into the other side of the SnapFast vector.
Function 2: Viral genomic insertion
We are designing a range of viral vectors containing PacI and AsiSI/NotI/NheI and SbfI sites. This will allow the direct cloning of your SnapFast expression cassettes into each of these viruses. The AsiSI and PacI sites are designed to be used to insert a gene (or concatemerised genes), including the promoters, into the viral vectors.
SwaI - ATTT'AAAT
The SwaI site has one function
Function: Change the origin of replication
We stock a range of bacterial origins of replication that can be used to change the copy number of the SnapFast plasmid in bacteria.
As with all our inserts that are flanked by two identical sites, we recommend using the closest possible unique restriction sites rather than the flanking sites. For example, to swap an origin of replication it may be easier to use PacI and AscI rather than SwaI, which is a blunt cutting enzyme. In the absence of unique sites, SwaI can be used but the cloning efficiency will be lower. We regularly swap and change the origins using the SwaI sites and find that although the efficiency of the ligation is lower, it very rarely fails to yield the correct clone. To determine the orientation of your insert we recommend sequencing it using one of our pre-designed primers (F1-10 and R1-10). Directional primers are also in development to enable PCR screening of each of our inserts that can be ligated in either orientation. However, the origins of replication that we sell will be fully functional independent of their orientation between the SwaI sites.
FseI - GGCCGG'CC
FseI site has one main function.
Important note: FseI is normally stored at -80°C and can lose activity quickly at -20°C. For optimal cloning, ensure that the FseI you are using is still fully active and add more than the protocol recommends!
Function: Insertion of reporter genes and mammalian selectable markers
Similar to the SbfI and AscI sites, the FseI site can be used to insert pre-designed secondary gene cassettes that encode reporter genes and mammalian selectable marker genes. This FseI site, and the AscI site, are the sites we recommend for these insertions if you do not need to transfer the genes into any viral vectors (which are currently in development). This is because these sites are outside of the PacI and AsiSI sites that are used to clone genes, or gene cassettes, into our viral vectors.
AscI - GG'CGCGCC
The AscI site has one main function.
Function 1: Insertion of reporter genes and mammalian selectable markers
Similar to the FseI and SbfI sites, the AscI site can be used to insert pre-designed secondary gene cassettes that encode reporter genes and mammalian selectable marker genes. This AscI site, and the FseI site, are the sites we recommend for these insertions if you do not need to transfer the pre-designed genes into any viral vectors (which are in development) after you have put them into the SnapFast vector. This is because these sites are outside of the PacI and AsiSI sites that are used to clone genes, or gene cassettes, into our viral vectors.
PmeI - GTTT'AAAC
The PmeI site has one function.
Function: Change the bacterial selectable marker
We stock a range of bacterial selectable markers that allow you to alter the antibiotic selection you use to grow your plasmids. This can help with recombination strategies and also to lower background contamination during ligations. For this reason all of our gene expression vectors, and vectors containing genes in the main MCS, are supplied in an ampicillin backbone, whilst all of our vectors that contain SnapFast compatible inserts are supplied in a Kanamycin resistant backbone.
As with all our inserts that are flanked by two identical sites, we recommend using the closest possible unique restriction sites rather than the paired sites that flank the insert. For example, to swap a bacterial selectable marker it may be easier to use AsiSI and AscI rather than PmeI, which is a blunt cutting enzyme. In the absence of unique sites, PmeI can be used but the cloning efficiency will be lower. We regularly swap and change the selection markers using the PmeI sites and although the ligation efficiency is generally lower, it almost always yields correct clones. To determine the orientation of your insert we recommend sequencing it using one of our pre-designed primers (F1-10 and R1-10). Directional primers are also in development to enable PCR screening of each of our inserts that can be ligated in either orientation. However, the antibiotic resistancce cassettes in our plasmids will function regardless of their orientation between the PmeI sites.
Materials
To continue reading please sign in or create an account.
Don't Have An Account?