T1660
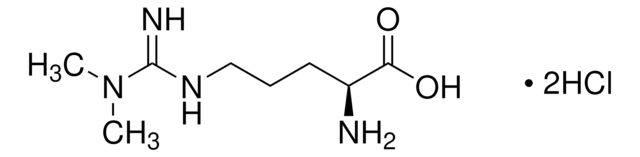
Nε,Nε,Nε-Trimethyllysine hydrochloride
≥97% (TLC)
Synonym(s):
1-Pentanaminium, 5-amino-5-carboxy-N,N,N-trimethyl-, chloride (1:1), (5S)-
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
Empirical Formula (Hill Notation):
C9H20N2O2 · HCl
CAS Number:
Molecular Weight:
224.73
MDL number:
UNSPSC Code:
12352202
PubChem Substance ID:
NACRES:
NA.26
Recommended Products
Quality Level
Assay
≥97% (TLC)
form
powder
contains
salts and water as balance
composition
Amino acid content, ~75%
technique(s)
LC/MS: suitable
color
white
storage temp.
−20°C
SMILES string
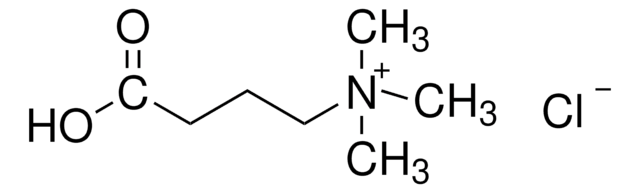
[Cl-].C[N+](C)(C)CCCC[C@H](N)C(O)=O
InChI
1S/C9H20N2O2.ClH/c1-11(2,3)7-5-4-6-8(10)9(12)13;/h8H,4-7,10H2,1-3H3;1H/t8-;/m0./s1
InChI key
ZKIJKCHCMFGMCM-QRPNPIFTSA-N
Related Categories
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point(F)
Not applicable
Flash Point(C)
Not applicable
Personal Protective Equipment
dust mask type N95 (US), Eyeshields, Gloves
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Andrew J Bannister et al.
The Journal of biological chemistry, 280(18), 17732-17736 (2005-03-12)
Methylation of lysine 4 of histone H3 (K4/H3) is linked to transcriptional activity, whereas methylation of K9/H3 is tightly associated with gene inactivity. These are well characterized sites of methylation within histones, but there are numerous other, less characterized, sites
Sara P Gaucher et al.
Journal of proteome research, 7(6), 2320-2331 (2008-04-18)
Recent developments in shotgun proteomics have enabled high-throughput studies of a variety of microorganisms at a proteome level and provide experimental validation for predicted open reading frames in the corresponding genome. More importantly, advances in mass spectrometric data analysis now
C J Rebouche et al.
The Journal of nutrition, 116(5), 751-759 (1986-05-01)
Rates of carnitine biosynthesis in mammals depend on the availability of substrates and the activity of enzymes subserving the pathway. This study was undertaken to test the hypothesis that the availability of epsilon-N-trimethyllysine is rate-limiting for synthesis of carnitine in
Jean-François Couture et al.
The Journal of biological chemistry, 281(28), 19280-19287 (2006-05-10)
SET domain enzymes represent a distinct family of protein lysine methyltransferases in eukaryotes. Recent studies have yielded significant insights into the structural basis of substrate recognition and the product specificities of these enzymes. However, the mechanism by which SET domain
Yasunori Tokuda et al.
Journal of bioscience and bioengineering, 111(4), 402-407 (2011-01-11)
The preparation of posttranslationally modified proteins is required to investigate the function and structure of modified proteins. However, homogeneously modified proteins are not easily isolated from natural sources or prepared using modification enzymes. Non-natural amino acid mutagenesis has enabled us
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service