10107921001
Roche
Nuclease S7
Micrococcal nuclease, from Staphylococcus aureus
Synonym(s):
s7 nuclease
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
UNSPSC Code:
12352204
Recommended Products
biological source
Staphylococcus aureus
Quality Level
form
lyophilized
specific activity
~15,000 U/mg
packaging
pkg of 15,000 U
manufacturer/tradename
Roche
optimum pH
7.5
shipped in
wet ice
Application
- Removal of endogenous RNA from in vitro translation systems
- RNA sequencing experiments
Nuclease S7 has been used in ribonuclease selection process for ribosome profiling.
Biochem/physiol Actions
Nuclease S7 is an endonuclease that mainly cleaves single-stranded substrates. It also cleaves double-stranded DNA or RNA. It cleaves the 5′-phosphodiester bonds of RNA and DNA to yield 3′-mononucleotides and oligonucleotides. Activity of the enzyme is dependent on Ca2+ and can be completely inactivated by the addition of EDTA. Nuclease S7 cleavage is highly specific to AT rich region than GC on the DNA.
Unit Definition
One unit releases a sufficient amount of acid-soluble oligonucleotides to produce an increase of 1.0 at A260.
Storage and Stability
Store unopened reagent at 2-8 °C; when diluted to 1 mg/mL, store at -15 to -25 °C.
Other Notes
For life science research only. Not for use in diagnostic procedures.
Signal Word
Danger
Hazard Statements
Precautionary Statements
Hazard Classifications
Eye Irrit. 2 - Resp. Sens. 1 - Skin Irrit. 2 - Skin Sens. 1 - STOT SE 3
Target Organs
Respiratory system
Storage Class Code
11 - Combustible Solids
WGK
WGK 1
Flash Point(F)
does not flash
Flash Point(C)
does not flash
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Organ-specific translation elongation rates measured by in vivo ribosome profiling.
Gerashchenko M V, et al.
bioRxiv, 279257 (2018)
High sequence specificity of micrococcal nuclease.
Dingwall C, et al.
Nucleic Acids Research, 9(12), 2659-2674 (1981)
Sequence specific cleavage of African green monkey ?-satellite DNA by micrococcal nuclease.
Horz W, et al.
Nucleic Acids Research, 11(13), 4275-4285 (1983)
Reimo Zoschke et al.
Proceedings of the National Academy of Sciences of the United States of America, 112(13), E1678-E1687 (2015-03-17)
Chloroplast genomes encode ∼ 37 proteins that integrate into the thylakoid membrane. The mechanisms that target these proteins to the membrane are largely unexplored. We used ribosome profiling to provide a comprehensive, high-resolution map of ribosome positions on chloroplast mRNAs
Ribonuclease selection for ribosome profiling.
Gerashchenko M V and Gladyshev V N
Nucleic Acids Research, 45(2), e6-e6 (2016)
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service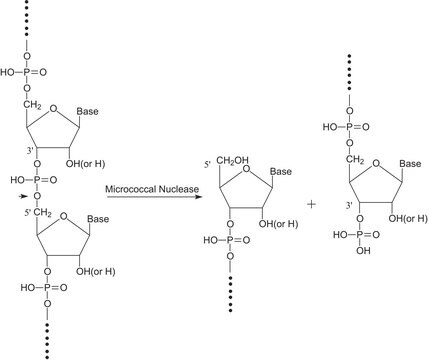









![Guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate ≥85% (HPLC), powder](/deepweb/assets/sigmaaldrich/product/structures/204/494/05808804-1ca7-44bf-b6c5-d4934dc7cb85/640/05808804-1ca7-44bf-b6c5-d4934dc7cb85.png)

