Determination of Eight Aminoglycosides in Pork Meat acc. to GB/T 21323-2007
Dean Duan, Senior Scientist
Merck R&D, APAC Lab, Shanghai, China
Abstract
Following the GB/T 21323-2007 method, an LC-MS/MS (ESI positive mode) method was developed to determine residues of eight aminoglycoside antibiotics, spectinomycin, hygromycin B, dihydrostreptomycin, amikacin, kanamycin, tobramycin, gentamicin and neomycin in pork meat. The cleanup of the pork meat sample was done by SPE using a Discovery® DSC-18 tube. An Ascentis® Express C18 UHPLC column was used to separate and quantify the aminoglycoside residues. The R2 values for the external calibration in the range of 1 to 5000 ng/mL were between 0.9927 and 0.9998 and the % recovery was 81.4 to 101.4%. The LOD and LOQ of the aminoglycosides in pork meat ranged from 0.2 to 0.8 µg/kg and 0.5 to 2.3 µg/kg respectively. The described method met the acceptance criteria of the official Chinese Method.
Section Overview
Introduction
Aminoglycosides are natural or semisynthetic antibiotics derived from actinomycetes.1 Most of aminoglycosides are widely used against bacteria and parasites in the production of pork, chicken, beef, milk, and eggs around the world, which could arouse side reactions and antimicrobial resistance to consumers (Schenck, 1998).2 Thus, governments around the world, and international regulatory bodies, including the European Union, the US Food and Drug Administration and the Codex Alimentarius are all intensifying their efforts to control veterinary antibiotics usage and issuing increasingly stringent regulations on maximum residue limits (MRLs).3,4 In order to protect public health, China has released national standard methods GB/T 21323-2007 for determination the residues of eight aminoglycosides (Figure 1) in food.5 Following this GB method, an application was developed for determination of eight aminoglycosides in pork meat. In this study the pork samples were cleaned-up by SPE using a Discovery® DSC-18 SPE tube and the resulting sample was run on an Ascentis® Express C18 UHPLC column to quantify the eight aminoglycosides by LC-MS/MS.
Figure 1.Structures of aminoglycosides determined.
Experimental
Standards and samples were prepared according to the following procedures:
Standard Preparation
- Weigh 4.28 g perfluorobutanoic acid and transfer it to a 1 L volumetric flask. Dilute to volume with water for a 20 mM perfluorobutanoic acid aqueous solution.
- Weigh 10 mg each of the eight aminoglycoside reference materials into individual 10 mL volumetric flasks (1 mg/ml).
- Add 7 mL methanol to each flask and sonicate for 5 mins. Top up to mark with methanol and mix well.
- Transfer 100 μL of each standard into a 10 mL volumetric flask and top up with 20 mM perfluorobutanoic acid (10 μg/mL).
- Dilute the 10 μg/mL standard to 1, 2, 5, 10, 20, 50, 100, 200, 500, 1000, 2000, and 5000 ng/mL with 20 mM perfluorobutanoic acid. Mix well before use.
Sample Preparation
- Phosphate buffer solution: Weigh 1.36 g of potassium dihydrogen phosphate, 0.15 g of Na2EDTA, and 20 g of trichloroacetic acid in a 1 L volumetric flask. Add 980 mL of water, adjust the pH to 4.0 using 1 M hydrochloric acid and make up to 1000 mL with water. This is phosphate buffer pH 4.0 containing 0.4 mM EDTA and 2% trichloroacetic acid.
- Homogenize pork samples with a high-speed tissue homogenizer.
- Weigh 5 g of homogenized pork sample into a 50 mL centrifuge tube and add 10 mL of phosphate buffer solution.
- Shake tubes for 10 min followed by centrifugation at 4500 rpm for 10 min.
- Transfer the supernatant to another 50 mL centrifuge tube.
- Repeat extraction with 10 ml phosphate buffer and pool the supernatant into one tube. Adjust pH to 3.5 ± 0.2 with 1.0 M HCl solution.
- Add 2.0 mL of 100 mM perfluorobutanoic acid aqueous solution and mix well for SPE clean-up.
- Perform SPE clean up according to the protocol in Table 1.
SPE Conditions |
|
|---|---|
Sample/matrix: | Pork sample extract |
SPE tube/cartridge: | Discovery® DSC-18, 500 mg/3 mL (52603-U) |
Conditioning: | 3 mL methanol, followed by 3 mL 20 mM perfluorobutanoic acid |
Sample Load: | Transfer the total collected extract volume and set a flow rate of ~1 drop/s |
Washing: | 3 mL of 20 mM perfluorobutanoic acid followed by 2 x 3 mL water. Drain for 5 min. |
Elution: | 5 mL of acetonitrile : 20 mM perfluorobutanoic acid (80:20 %v/v) |
Eluate post-treatment: | Evaporate to dryness under nitrogen at 40 °C and reconstitute to 1 mL 20 mM perfluorobutanoic acid. |
LC-MS/MS Analysis
Standards and samples were analyzed by LC-MS using the method described in Table 2.
LC Conditions |
| ||||
|---|---|---|---|---|---|
Instrument: | Acquity UPLC I-class plus | ||||
Column: | Ascentis® Express C18 UHPLC, 2 µm, 100 x 2.1 mm I.D. (50813-U) | ||||
Mobile phase: | [A] Water with 20 mM perfluorobutanoic acid [B] Acetonitrile/water (80/20, v/v) with 20 mM perfluorobutanoic acid | ||||
Gradient: | Time (min) | A% | B% | ||
0 | 85 | 15 | |||
1.5 | 85 | 15 | |||
7.5 | 25 | 75 | |||
8.9 | 25 | 75 | |||
8.91 | 85 | 15 | |||
10.0 | 85 | 15 | |||
Flow rate: | 0.3 mL/min | ||||
Pressure: | 5441 to 6913 psi | ||||
Column temp.: | 25℃ | ||||
Detector: | MS, MRM settings see Table 3 | ||||
Injection: | 10 μL | ||||
Samples: | Prepared as described | ||||
MS Conditions |
|
| |||
Instrument: | Xevo TQ-S |
| |||
Polarity: | Positive and negative |
| |||
Spray voltage: | 3.0 kV (ESI+) |
| |||
Capillary temp: | 500℃ |
| |||
Desolvation (L/Hr): | 1000 |
| |||
Cone (L/Hr): | 150 |
| |||
Nebulizer (Bar): | 7 |
| |||
Transition (m/z) | Cone (V) | Collision (V) | Dwell (s) | |
|---|---|---|---|---|
Spectinomycin | 333.1 -> 98.1 | 60 | 26 | 0.025 |
Hygromycin B | 528.3 -> 177.1 | 54 | 28 | 0.025 |
Dihydrostreptomycin | 584.3 -> 263.2 | 52 | 22 | 0.025 |
Amikacin | 586.3 -> 425.2 | 54 | 18 | 0.025 |
Kanamycin | 485.4 -> 163.2 | 66 | 24 | 0.025 |
Tobramycin | 468.4 -> 205.2 | 32 | 19 | 0.025 |
Gentamicin | 478.3 -> 157.3 | 34 | 22 | 0.025 |
Neomycin | 615.4 -> 293.1 | 62 | 24 | 0.025 |
Method & System Suitability
Acceptance Criteria as described in GB/T 21323-2007
- Critical ions must be present and exceed a signal-to-noise ratio > 5.
- Linear correlation coefficient: R2 > 0.99
- Aminoglycoside recovery with extraction and cleanup by SPE: 70–120%.
- For neomycin, hygromycin B, LOQ < 100 mg/kg; for other aminoglycosides, LOQ < 20 mg/kg.
Results & Discussion
Chromatographic results for a standard, blank and spiked pork meat samples are displayed in the Figures 2-4. The retention times for the 200 ng/mL standard solution are shown in Table 4. The linearity and derived sensitivity for a pork meat sample can be seen in Tables 5. All compounds showed R2 values above 0.99. The repeatability for the injection of the 200 ng/mL standard ranged from 1.72 to 3.29 % RSD (Table 6). All critical product ions are presented with signal-to-noise ratio (S/N) >5 during analysis. As example the calibration curve for spectinomycin is displayed in Figure 5. The method recovery for the 8 aminoglycosides spiked at 50 μg/kg into pork meat after SPE cleanup ranged from 81.4 to 101.4% (Table 7). The shown method met all acceptance criteria stated by the GB/T 21323-2007 method.
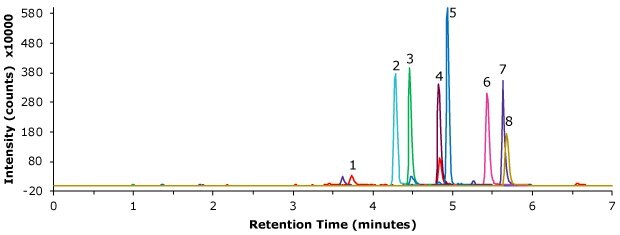
Figure 2.Injection of an aminoglycoside standard, 200 ng/mL (MRM product ion traces).
Peak ID | Compound | Retention Time (min) |
|---|---|---|
1 | Spectinomycin | 3.69 |
2 | Hygromycin B | 4.27 |
3 | Dihydrostreptomycin | 4.45 |
4 | Amikacin | 4.80 |
5 | Kanamycin | 4.90 |
6 | Tobramycin | 5.42 |
7 | Gentamicin | 5.60 |
8 | Neomycin | 5.65 |

Figure 3.Injection of a blank pork meat sample.

Figure 4.Analysis of aminoglycosides in pork meat spiked at 100 μg/kg after SPE cleanup.
Compound | Calibration (ng/mL) | No. of Calibrators | R2 | LOD (µg/kg) | LOQ (µg/kg)* |
|---|---|---|---|---|---|
Spectinomycin | 1 - 5000 | 12 | 0.9927 | 0.8 | 2.3 |
Hygromycin B | 0.9963 | 0.2 | 0.5 | ||
Dihydrostreptomycin | 0.9984 | 0.3 | 0.9 | ||
Amikacin | 0.9976 | 0.2 | 0.6 | ||
Kanamycin | 0.9998 | 0.3 | 0.9 | ||
Tobramycin | 0.9970 | 0.2 | 0.6 | ||
Gentamicin | 0.9991 | 0.4 | 1.3 | ||
Neomycin | 0.9986 | 0.3 | 1.0 | ||
*Note: Acceptance criteria for sensitivity by the GB method are for neomycin and hygromycin B an LOQ <100 mg/kg; for the other aminoglycosides an LOQ of <20 mg/kg | |||||
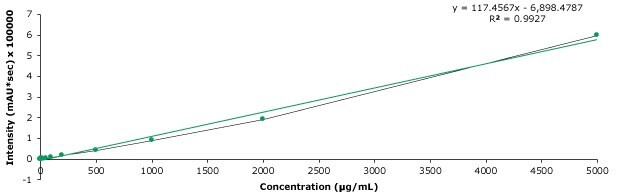
Figure 5.Calibration curve for spectinomycin.
Compound | Mean Area | Std Dev | RSD (%) |
|---|---|---|---|
Spectinomycin | 18469 | 318 | 1.72 |
Hygromycin B | 177800 | 4392 | 2.47 |
Dihydrostreptomycin | 171061 | 3927 | 2.30 |
Amikacin | 148945 | 3851 | 2.59 |
Kanamycin | 257395 | 6095 | 2.37 |
Tobramycin | 143749 | 4728 | 3.29 |
Gentamycin | 152902 | 4405 | 2.88 |
Neomycin | 87728 | 2127 | 2.42 |
Compound | Sample 1 (%) | Sample 2 (%) | Sample 3 (%) | Mean | SD | RSD (%) |
|---|---|---|---|---|---|---|
Spectinomycin | 96.5 | 90 | 86.8 | 91.1 | 4.9 | 5.4 |
Hygromycin | 87.9 | 88.5 | 87.9 | 88.1 | 0.3 | 0.4 |
Dihydrostreptomycin | 81 | 86.2 | 84.2 | 83.8 | 2.6 | 3.1 |
Amikacin | 81.8 | 84.2 | 78.2 | 81.4 | 3.0 | 3.7 |
Kanamycin | 91.4 | 94.2 | 88.9 | 91.5 | 2.7 | 2.9 |
Tobramycin | 102.2 | 102.7 | 99.4 | 101.4 | 1.8 | 1.8 |
Gentamicin | 84.9 | 84.4 | 78.2 | 82.5 | 3.7 | 4.5 |
Neomycin | 83.2 | 85.6 | 79.3 | 82.7 | 3.2 | 3.8 |
Conclusion
In this determination of eight aminoglycoside residues in pork meat following the GB/T 21323 -2007 method, the SPE cleanup was performed with a Discovery® DSC-18 SPE tube, and for the following LC-MS/MS analysis an Ascentis® Express C18 UHPLC column with 2 µm particles was used. The developed method showed good reproducibility, sensitivity, and linearity over a wide concentration range and met the acceptance criteria as described in GB/T 21323-2007. The R2 for the eight aminoglycoside residues from 1 to 5000 ng/mL ranged from 0.9927 to 0.9998. The calibration curve derived LODs and LOQs ranged from 0.2 to 0.8 μg/kg and 0.5 to 2.3 μg/kg respectively. The % recovery of the aminoglycosides for a 50 µg/kg spiked sample after SPE cleanup ranged from 81.4 to 101.4%.
See more applications for Food & Beverage testing.
References
To continue reading please sign in or create an account.
Don't Have An Account?