Polishing of MAbs Using Capto Adhere ImpRes in Bind/Elute Mode
In these studies, the binding capacity for MAbs and the efficiency in the clearance of impurities using Capto adhere ImpRes in bind/elute mode was evaluated. The studies present results from optimization of the loading conditions using the DoE approach. The effects of buffer, pH, conductivity, and sample load were investigated. The studies include measurement of static and dynamic binding capacities (SBC and DBC, respectively) at various binding conditions, as well as screening and optimization of gradient- and step-elution conditions.
Two different MAbs were studied. The results showed high yields of monomeric MAb, as well as good clearance of aggregates, HCP, and leached protein A.
Materials and Methods
Start Material
The two MAbs used in this study were initially purified from CHO cell supernatant by protein A affinity chromatography (Table 1).
Determination of SBC
SBC was determined in 6 μL PreDictor 96-well filter plates. Equilibration of wells in the filter plates was performed by addition of 200 μL of loading buffer per well followed by agitation at 1100 rpm for 1 min, after which the buffer was removed by vacuum extraction. The equilibration step was performed three times. MAb solution (200 μL volume, 4 mg/mL sample load, corresponding to 133 mg MAb/mL chromatography medium) was added to each well followed by agitation for 90 min. Unbound material (flowthrough fraction) was removed by centrifugation for 3 min, and MAb concentration was determined by measurement of absorbance at 280 nm. SBC was calculated according to:

where Vload = load volume, Vmedium = medium volume in each well, Cini = MAb concentration in the sample, and CFT = MAb concentration in the flowthrough fraction.
Determination of DBC
DBC was determined by frontal analysis using an ÄKTA chromatography system. The UV absorbance at 280 nm was used for determination of breakthrough. Before frontal analysis, the MAb solution was injected bypassing the column to obtain a maximum absorbance value. DBC was then calculated according to:
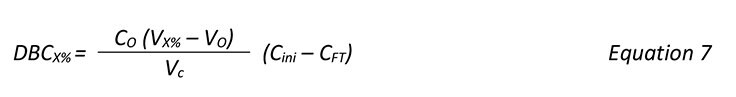
where C0 = MAb concentration in the sample (mg/mL), VX% = load volume (mL) at x% breakthrough,
V0 = void volume (mL), and Vc = volumetric bed volume (mL).
Screening of Elution Conditions
Measurement of yield at different elution conditions was performed in PreDictor 96-well filter plates. Equilibration of wells in the filter plates was performed by the addition of 200 μL of loading buffer per well followed by agitation at 1100 rpm for 1 min, after which the buffer was removed by centrifugation. The equilibration step was performed three times. MAb solution (200 μL, 2.8 mg/mL, corresponding to 93 mg MAb/mL medium) was added to each well followed by agitation for 60 min. Unbound material was removed by centrifugation. Elution of bound material was then performed by the addition of 200 μL of elution buffer/well; the elution step was performed three times. Mab concentration was determined by measurement of absorbance at 280 nm.
The yield was calculated according to:

where Veluate = eluate volume, Celuate 1, 2, 3 = MAb concentration in eluates 1 to 3,
Vload = load volume, and Cini = MAb concentration in MAb solution.
Optimization of Step Elution Conditions
Conditions for step elution were investigated in a packed column using ÄKTA pure chromatography system, DoE, and scouting functionalities included in UNICORN™ 6.3.
Determination of Aggregates and Aggregate Clearance
Fractions from the chromatographic runs were collected and analyzed by analytical GF on a Superdex 200 5/150 GL column. The peaks were integrated, and the D/A concentrations (in percent) were estimated. The cumulated yield of monomers was plotted against cumulated aggregates (Figure 1).
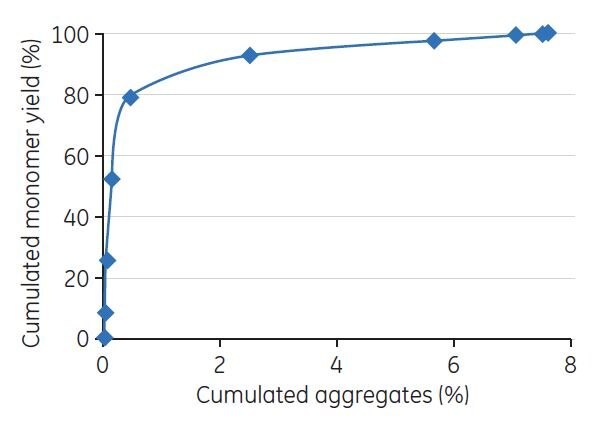
Figure 1.Evaluation of gradient elution was performed by GF. The figure shows an example of the resulting plot of cumulated yield of monomers vs cumulated aggregates derived from the GF analysis.
Protein A and HCP ELISA
The protein A concentration in the start materials and flowthrough fractions was determined by Protein A ELISA kit (Repligen). HCP concentration was determined by HCP ELISA (Cygnus Technologies).
Results and Discussion
Case Study, MAb A
The case study with MAb A shows a suggested workflow for method development including screening of conditions for SBC and DBC, screening of elution conditions, and optimization of conditions for step elution.
SBC
To find optimal binding capacity for MAb A, SBC was determined in 6 μL PreDictor 96-well filter plates. Binding pH was varied between pH 4.0 and 8.02,3 and the salt concentration from 0 to 500 mM NaCl. All samples and buffers were prepared automatically using a Tecan robot. The results show that the highest SBC was obtained at high pH and low salt concentration (Figure 2., orange region). Based on these results, a narrower range of pH and NaCl concentration was used for further investigation of conditions for DBC.
2 Binding buffers were citrate, pH 4; acetate, pH 4.6 and 5.7; phosphate, pH 5.7, 6.3, and 6.9; and Tris, pH 7.4 and 8.0. The ionic strength from the buffer salts was kept constant at 40 mM.
3 To avoid deamidation of the MAb, pH should normally be maintained below pH 8.0.
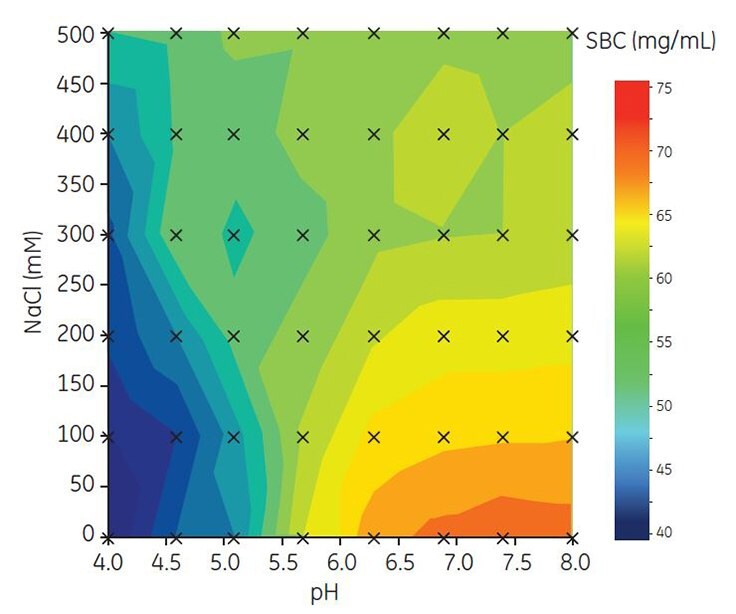
Figure 2. Contour map showing screening of SBC for Capto adhere ImpRes.
DBC
The influence of pH and salt concentration on DBC was measured by DoE using Capto adhere ImpRes packed in a Tricorn 5/50 column. Based on the results for SBC, binding pH was varied between pH 6.0 and 7.84 and salt concentration from 0 to 200 mM NaCl. In addition, the residence time was varied from 2 to 8 min.
The results from the DoE are shown in Figure 3. Modeling of data was performed using MODDE v9.0 software, resulting in a good model fit and predictive power (data not shown). In accordance with the trend for SBC, an increase in pH and decrease in salt concentration resulted in higher DBC, while lower capacity was obtained at short residence time. Further experiments described below were performed using binding with 40 mM sodium phosphate, pH 7.8.
4 Binding buffers: Sodium phosphate, 0 to 200 mM NaCl, pH 6 to 7.8. The ionic strength from the buffer salts was kept constant at 110 mM.
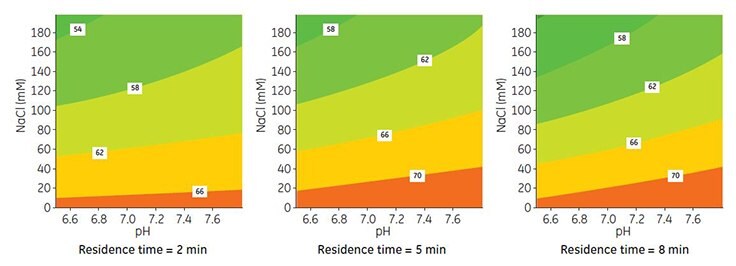
Figure 3. Contour maps for DBC at 10% breakthrough and different residence times on Capto adhere ImpRes
Screening of Elution Conditions
Measurement of yield at different elution conditions was performed in 96-well filter plates as described in “Materials and methods.” Binding was performed in 40 mM sodium phosphate, pH 7.8. Elution pH was varied between 4.5 and 8.0 and salt concentration between 0 and 1 M NaCl. The result, Figure 4, shows that the highest yield was obtained at low pH and low salt concentration. Based on this result, further studies of elution conditions were performed by gradient elution in packed columns.

Figure 4. Screening of elution conditions in PreDictor plate. Yield obtained by varying elution pH between 4.5 and 8.0 and NaCl concentration between 0 and 1 M. Evaluation performed by Assist software for PreDictor plates.
Gradient Elution
Gradient elution was performed from 40 mM sodium phosphate, pH 7.8 to 20 mM sodium phosphate, 20 mM citrate, pH 4.0 with or without the addition of 100 mM NaCl5. Chromatograms are shown in Figure 5. Fractions were collected and analyzed by GF. The cumulated concentration of aggregates (%) vs cumulated yield of monomeric MAb (%) was calculated according to “Materials and methods.” The results showed that the addition of 100 mM NaCl in the elution buffer resulted in slightly lower elution pH, lower aggregate content, and a broader elution peak than elution buffer without NaCl (Table 2).
5 A mixed buffer with ionic strength that is too high might result in elution of MAb during the wash step or early in the gradient.
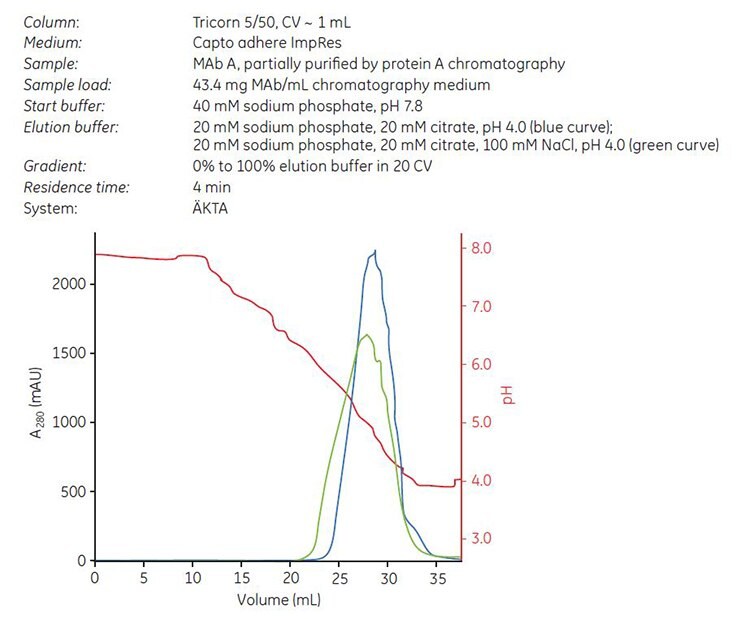
Figure 5. Gradient elution on Capto adhere ImpRes using elution buffer with NaCl (green curve) and without NaCl (blue curve) of MAb A, which was partially purified by protein A affinity chromatography.
Step Elution
Based on results from screening in 96-well filter plates and gradient elution, conditions for step elution were further investigated in a packed column using DoE, varying sample load between
approximately 50% and 70% of DBC (37.2 to 49.6 mg MAb/mL chromatography medium). Elution pH was varied between 3.5 and 4.5, and salt concentration between 0 and 100 mM NaCl. The responses from the design were yield, aggregate concentration, pool volume, HCP, and protein A concentration. The results from the design are shown in Table 3.
Modeling of the experimental data was performed with MODDE v9.0 software. Good models were obtained for all responses except for protein A6. The model showed that the only significant factor was elution pH. Thus, a higher elution pH resulted in lower yield, lower aggregate concentration, higher pool volume, and lower HCP concentration (Figure 6).
6 As the values and the variation of protein A concentration in the elution pools were very low, no model could be obtained for this response.
1 ND = Not determined.
2 LOQ = Limit of quantitation
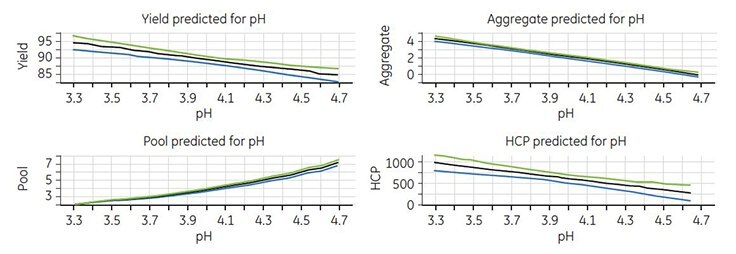
Figure 6.Response plots for yield, pool volume, aggregate, and HCP concentrations.
Verification of the Design
The model suggested an elution pH of 4.5 (0 M NaCl) and a sample load of 70% of DBC (≈ 50 mg/mL).
Column verification of the method was performed on a Tricorn 5/50 column. The obtained result was in good agreement with the expected result for yield, pool volume, and aggregate and HCP clearance (Table 4). The relatively high initial HCP level in the sample used accounts for the high HCP level after polishing. HCP levels could be further reduced, either by including a wash step before elution of the MAb or by addition of a third purification step.
Case Study, MAb B
The related multimodal anion exchanger, Capto adhere, has been successful for MAb polishing in flowthrough mode. However, Capto adhere has also found use in bind/elute mode, even though the particle size is not optimal. In a case study using MAb B, the performance of Capto adhere in bind/elute mode was compared with that of Capto adhere ImpRes, considering DBC at various residence times and gradient and step-elution conditions.
SBC and DBC
SBC and DBC for MAb B were determined using the same methodology as shown in the first case study. Highest SBC and DBC were obtained at high pH and low ionic strength (i.e., 20 mM sodium phosphate, pH 7.87).
7 To avoid deamidation of the MAb, pH should normally be maintained below pH 8.0.
DBC VS Residence Time
DBC at 10% breakthrough for Capto adhere ImpRes and Capto adhere was measured at different residence times (linear flow rates) in the range of 1 to 10 min. As seen in Figure 7, DBC for Capto adhere ImpRes is higher and less sensitive to residence time than is the case for Capto adhere. Capto adhere ImpRes can therefore be operated at shorter residence times (i.e., higher flow rates) while maintaining process robustness with regard to capacity8.
8 Due to pressure-flow limitations, a maximum bed height of 10 cm is recommended at 2 min residence time.
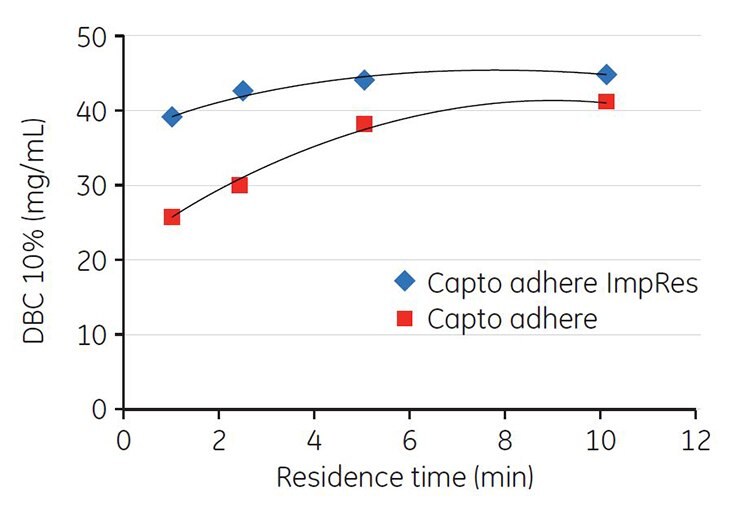
Figure 7.DBC vs residence time. DBC at 10% breakthrough measured in 28 mM sodium phosphate, pH 7.75.
Gradient Elution
Gradient elution by pH was performed on Capto adhere ImpRes. Unlike the example with Mab A, the addition of NaCl to the elution buffer resulted in a narrower elution peak (Figure 8, green curve). Collected fractions were analyzed by GF and the cumulated yield of monomer was plotted against the cumulated concentration of aggregates. The result shows a good separation between monomer and aggregates, and that separation was improved on Capto adhere ImpRes compared with Capto adhere (Figure 9).
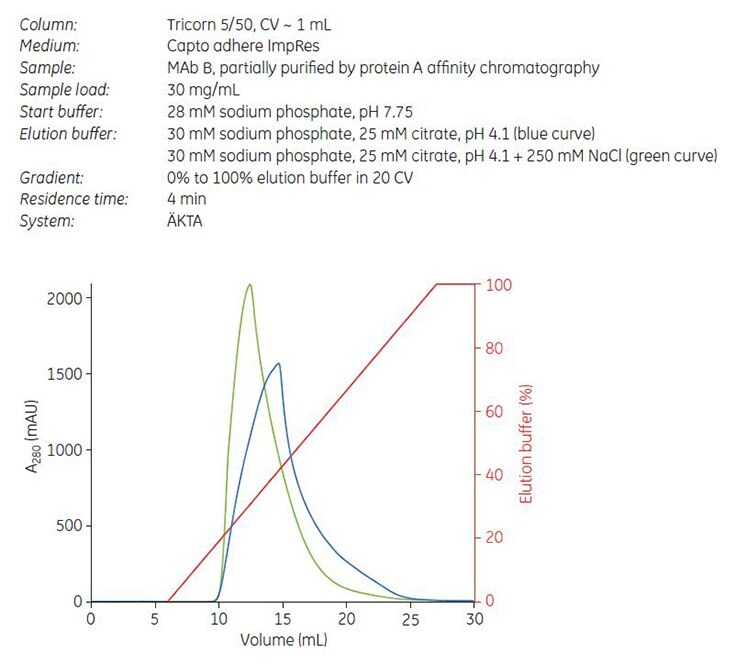
Figure 8.Gradient elution of MAb B from Capto adhere ImpRes with (green curve) and without (blue curve) NaCl in elution buffer.
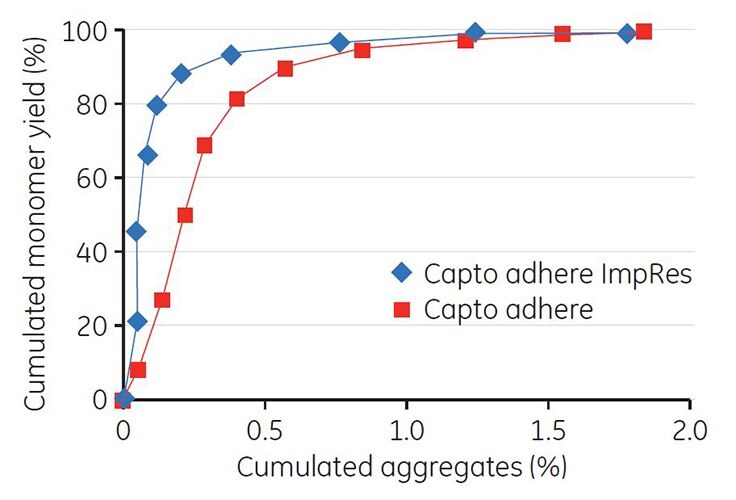
Figure 9.Cumulated aggregates vs cumulated MAb monomer yield after gradient elution using Capto adhere ImpRes and Capto adhere.
Step Elution
From the gradient elution results above, step elution from Capto adhere ImpRes and Capto adhere was performed at pH 6.5 and 62.5 mM NaCl (i.e., 25% of elution buffer, Figure 10). The sample load was 70% of DBC 10%. Fractions were pooled and analyzed for yield, aggregate, and HCP concentration. Despite 20% higher load, step elution from Capto adhere ImpRes resulted in higher yield and improved aggregate clearance compared with Capto adhere (Table 5). HCP levels were below the detection limit for ELISA.
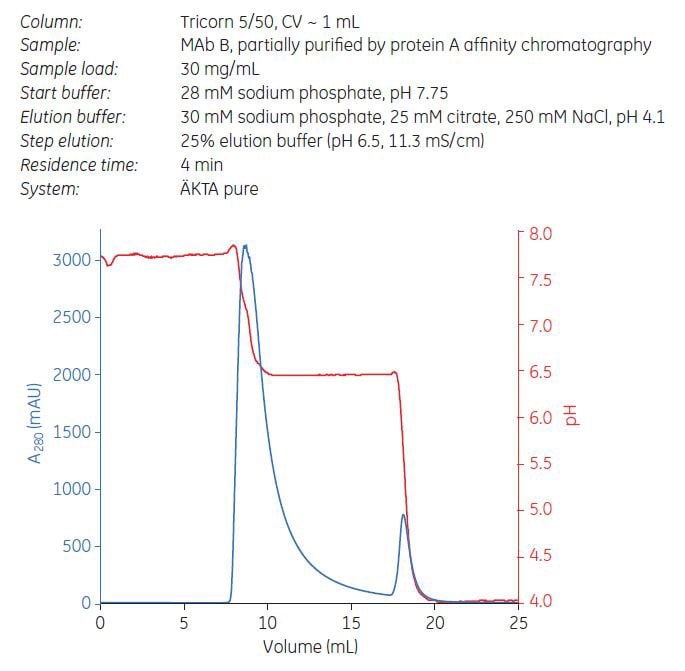
Figure 10.Step elution of MAb B using Capto adhere ImpRes.
Conclusions
In this work, results have been presented from two case studies using Capto adhere ImpRes, a multimodal anion exchanger designed for polishing. Two different MAbs were purified in bind/elute mode. The results show high yields of MAb monomers and good clearance of aggregates, HCP, and leached protein A.
To continue reading please sign in or create an account.
Don't Have An Account?