908401
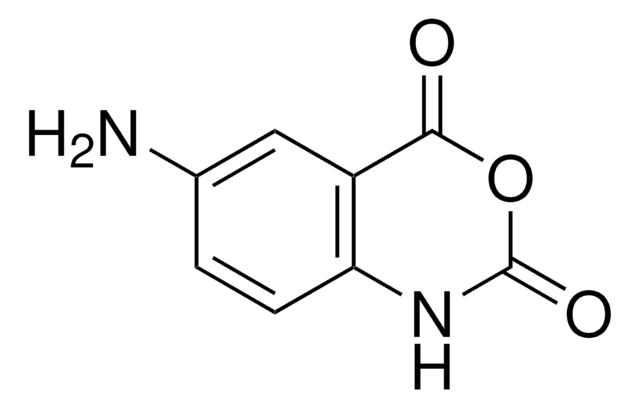
1-Methyl-7-nitroisatoic anhydride
동의어(들):
1-Methyl-7-nitro-2H-3,1-benzoxazine-2,4(1H)-dione, 1-methyl-7-nitro-2H-3,1-Benzoxazine-2,4(1H)-dione, 1M7, RNA SHAPE probe, Reagent for RNA SHAPE-MaP
로그인조직 및 계약 가격 보기
모든 사진(3)
About This Item
실험식(Hill 표기법):
C9H6N2O5
CAS Number:
Molecular Weight:
222.15
MDL number:
UNSPSC 코드:
12352119
NACRES:
NA.22
추천 제품
양식
powder
mp
204.5 °C
저장 온도
2-8°C
SMILES string
[N+](=O)([O-])c1cc2[n]([c]([o][c](c2cc1)=O)=O)C
InChI
1S/C9H6N2O5/c1-10-7-4-5(11(14)15)2-3-6(7)8(12)16-9(10)13/h2-4H,1H3
InChI key
MULNCJWAVSDEKJ-UHFFFAOYSA-N
애플리케이션
1-Methyl-7-nitroisatoic anhydride (1M7) is used as an in vivo SHAPE-MaP reagent for live cell RNA structure analysis at single nucleotide resolution. SHAPE -- or selective 2′-hydroxyl acylation analyzed by primer extension -- uses small, electrophilic chemical probes such as 1M7 to react with the 2′-hydroxyl group and provides insight to RNA structure. When combined with mutational profiling (MaP), quantitative nucleotide measurements are possible for entire transciptomes. Together, these methods deepen the understanding of RNA interactions and regions that may be exploited for design of RNA-targeting small-molecule drugs.
기타 정보
Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing
SnapShot: RNA Structure Probing Technologies
Detection of RNA-Protein Interactions in Living Cells with SHAPE
Standardization of RNA Chemical Mapping Experiments
In-cell RNA structure probing with SHAPE-MaP
A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry
SnapShot: RNA Structure Probing Technologies
Detection of RNA-Protein Interactions in Living Cells with SHAPE
Standardization of RNA Chemical Mapping Experiments
In-cell RNA structure probing with SHAPE-MaP
A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry
관련 제품
제품 번호
설명
가격
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
Flash Point (°F)
Not applicable
Flash Point (°C)
Not applicable
Katherine E Deigan et al.
Proceedings of the National Academy of Sciences of the United States of America, 106(1), 97-102 (2008-12-26)
Almost all RNAs can fold to form extensive base-paired secondary structures. Many of these structures then modulate numerous fundamental elements of gene expression. Deducing these structure-function relationships requires that it be possible to predict RNA secondary structures accurately. However, RNA
Scott P Hennelly et al.
Nucleic acids research, 39(6), 2416-2431 (2010-11-26)
Riboswitches are non-coding RNAs that control gene expression by sensing small molecules through changes in secondary structure. While secondary structure and ligand interactions are thought to control switching, the exact mechanism of control is unknown. Using a novel two-piece assay
A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry.
Stefanie A Mortimer et al.
Journal of the American Chemical Society, 129(14), 4144-4145 (2007-03-21)
Anthony M Mustoe et al.
Cell, 173(1), 181-195 (2018-03-20)
mRNAs can fold into complex structures that regulate gene expression. Resolving such structures de novo has remained challenging and has limited our understanding of the prevalence and functions of mRNA structure. We use SHAPE-MaP experiments in living E. coli cells to
Matthew J Smola et al.
Biochemistry, 54(46), 6867-6875 (2015-11-07)
SHAPE-MaP is unique among RNA structure probing strategies in that it both measures flexibility at single-nucleotide resolution and quantifies the uncertainties in these measurements. We report a straightforward analytical framework that incorporates these uncertainties to allow detection of RNA structural
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.

![1,4-Diazabicyclo[2.2.2]octane ReagentPlus®, ≥99%](/deepweb/assets/sigmaaldrich/product/structures/366/129/a6ff4175-974d-4fac-9038-b35e508ef252/640/a6ff4175-974d-4fac-9038-b35e508ef252.png)
![1,8-Diazabicyclo[5.4.0]undec-7-ene 98%](/deepweb/assets/sigmaaldrich/product/structures/120/564/5b373e23-1624-489c-8efb-692de0f96ffb/640/5b373e23-1624-489c-8efb-692de0f96ffb.png)