920800
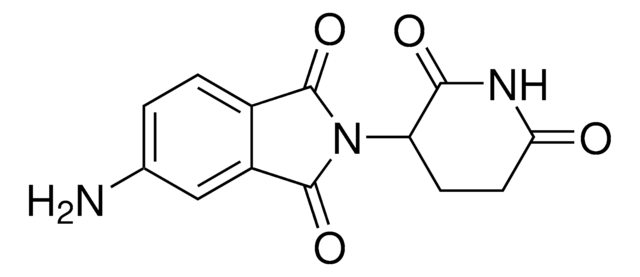
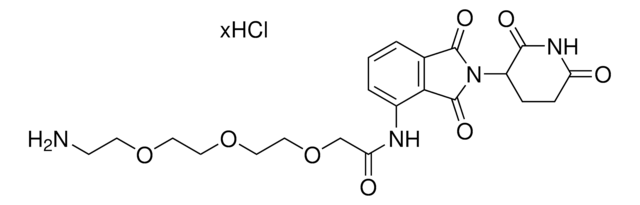
C5 Lenalidomide-methylamino-PEG1-NH2 hydrochloride
≥95%
동의어(들):
3-((2-(2-Aminoethoxy)ethyl)(methyl)amino)-N-(2-(2,6-dioxopiperidin-3-yl)-1-oxoisoindolin-5-yl)propanamide hydrochloride, Crosslinker−E3 Ligase ligand conjugate, Protein degrader building block for PROTAC® research, Template for synthesis of targeted protein degrader
로그인조직 및 계약 가격 보기
모든 사진(1)
About This Item
실험식(Hill 표기법):
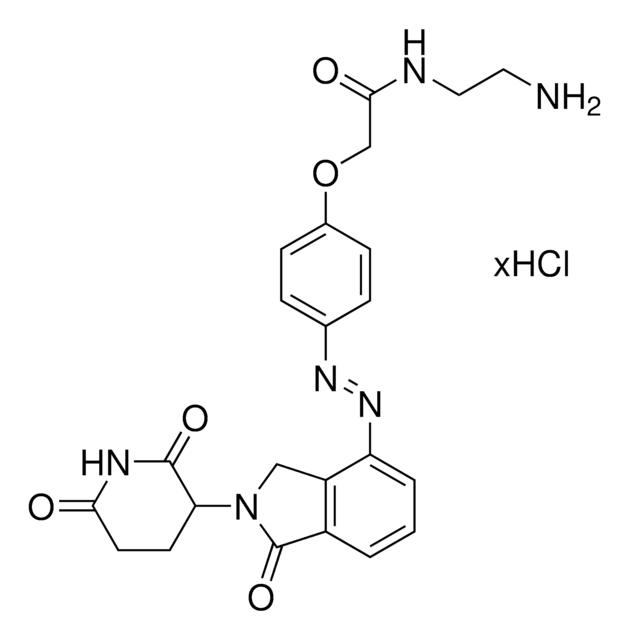
C21H29N5O5 · xHCl
Molecular Weight:
431.49 (free base basis)
UNSPSC 코드:
12352101
NACRES:
NA.22
추천 제품
ligand
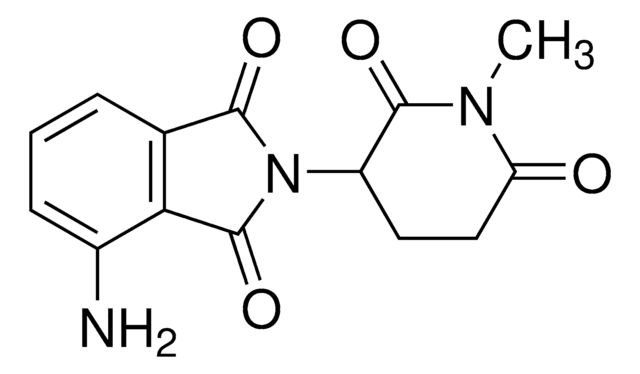
C5 Lenalidomide
Quality Level
분석
≥95%
형태
(Powder or Crystal or Solid or Chunks)
반응 적합성
reactivity: carboxyl reactive
reagent type: ligand-linker conjugate
작용기
amine
저장 온도
2-8°C
SMILES string
O=C1N(C2CCC(NC2=O)=O)CC3=CC(NC(CCN(C)CCOCCN)=O)=CC=C31.Cl
애플리케이션
Protein degrader building block C5 Lenalidomide-methylamino-PEG1-NH2 hydrochloride enables the synthesis of molecules for targeted protein degradation and PROTAC (proteolysis-targeting chimeras) technology. This conjugate contains a Cereblon (CRBN)-recruiting ligand with alternative exit vector, a linker, and a pendant amine for reactivity with a carboxylic acid on the target ligand. Because even slight alterations in ligands and crosslinkers can affect ternary complex formation between the target, E3 ligase, and PROTAC, many analogs are prepared to screen for optimal target degradation. When used with other protein degrader building blocks with a pendant amine, parallel synthesis can be used to more quickly generate PROTAC libraries that feature variation in crosslinker length, composition, and E3 ligase ligand.
Automate your CRBN-PEG based PROTACs with Synple Automated Synthesis Platform (SYNPLE-SC002)
Automate your CRBN-PEG based PROTACs with Synple Automated Synthesis Platform (SYNPLE-SC002)
기타 정보
Technology Spotlight: Degrader Building Blocks for Targeted Protein Degradation
Portal: Building PROTAC® Degraders for Targeted Protein Degradation
Targeted Protein Degradation by Small Molecules
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Impact of linker length on the activity of PROTACs
Portal: Building PROTAC® Degraders for Targeted Protein Degradation
Targeted Protein Degradation by Small Molecules
Small-Molecule PROTACS: New Approaches to Protein Degradation
Targeted Protein Degradation: from Chemical Biology to Drug Discovery
Impact of linker length on the activity of PROTACs
법적 정보
PROTAC is a registered trademark of Arvinas Operations, Inc., and is used under license
관련 제품
제품 번호
설명
가격
Storage Class Code
11 - Combustible Solids
WGK
WGK 3
가장 최신 버전 중 하나를 선택하세요:
Daniel P Bondeson et al.
Annual review of pharmacology and toxicology, 57, 107-123 (2016-10-13)
Protein homeostasis networks are highly regulated systems responsible for maintaining the health and productivity of cells. Whereas therapeutics have been developed to disrupt protein homeostasis, more recently identified techniques have been used to repurpose homeostatic networks to effect degradation of
Kedra Cyrus et al.
Molecular bioSystems, 7(2), 359-364 (2010-10-06)
Conventional genetic approaches have provided a powerful tool in the study of proteins. However, these techniques often preclude selective manipulation of temporal and spatial protein functions, which is crucial for the investigation of dynamic cellular processes. To overcome these limitations
Momar Toure et al.
Angewandte Chemie (International ed. in English), 55(6), 1966-1973 (2016-01-13)
The current inhibitor-based approach to therapeutics has inherent limitations owing to its occupancy-based model: 1) there is a need to maintain high systemic exposure to ensure sufficient in vivo inhibition, 2) high in vivo concentrations bring potential for off-target side effects, and 3) there is
Philipp M Cromm et al.
Cell chemical biology, 24(9), 1181-1190 (2017-06-27)
Traditional pharmaceutical drug discovery is almost exclusively focused on directly controlling protein activity to cure diseases. Modulators of protein activity, especially inhibitors, are developed and applied at high concentration to achieve maximal effects. Thereby, reduced bioavailability and off-target effects can
자사의 과학자팀은 생명 과학, 재료 과학, 화학 합성, 크로마토그래피, 분석 및 기타 많은 영역을 포함한 모든 과학 분야에 경험이 있습니다..
고객지원팀으로 연락바랍니다.