A9934
Aminopeptidase I from Streptomyces griseus
lyophilized powder, ≥200 units/mg protein
Synonym(s):
Leucine Aminopeptidase IV
About This Item
Recommended Products
form
lyophilized powder
Quality Level
specific activity
≥200 units/mg protein
mol wt
21 kDa by gel filtration
33 kDa by SDS-PAGE
composition
Protein, 40-60% Lowry
storage temp.
−20°C
Related Categories
General description
Application
- to test the biochar exposure effect on the enzyme activity
- in circular dichroism (CD) spectroscopy studies
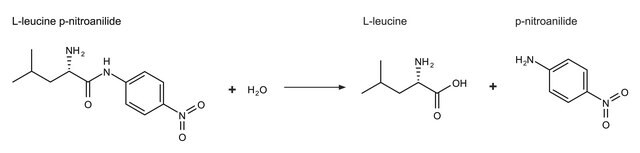
- as a positive control in p-nitroanilide degradation assay
Biochem/physiol Actions
Packaging
Unit Definition
Physical form
Preparation Note
Other Notes
signalword
Danger
hcodes
Hazard Classifications
Eye Irrit. 2 - Resp. Sens. 1 - Skin Irrit. 2 - STOT SE 3
target_organs
Respiratory system
Storage Class
11 - Combustible Solids
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
ppe
Eyeshields, Gloves, type N95 (US)
Choose from one of the most recent versions:
Certificates of Analysis (COA)
Don't see the Right Version?
If you require a particular version, you can look up a specific certificate by the Lot or Batch number.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service