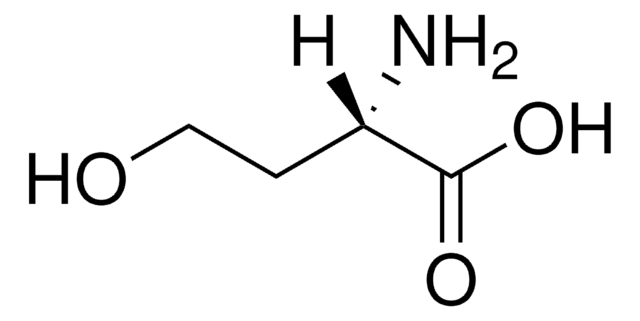
E1260
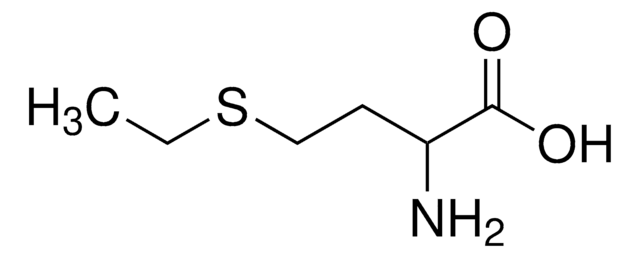
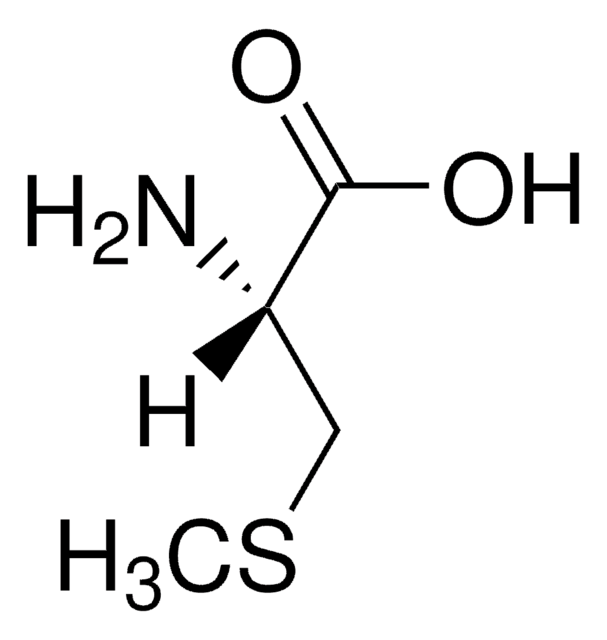
L-Ethionine
≥99% (TLC)
Synonym(s):
L-2-Amino-4-(ethylthio)butyric acid
About This Item
Recommended Products
product name
L-Ethionine, ≥99% (TLC)
assay
≥99% (TLC)
form
powder
color
white
mp
280 °C (dec.) (lit.)
storage temp.
−20°C
SMILES string
CCSCC[C@H](N)C(O)=O
InChI
1S/C6H13NO2S/c1-2-10-4-3-5(7)6(8)9/h5H,2-4,7H2,1H3,(H,8,9)/t5-/m0/s1
InChI key
GGLZPLKKBSSKCX-YFKPBYRVSA-N
Looking for similar products? Visit Product Comparison Guide
Biochem/physiol Actions
signalword
Warning
hcodes
Hazard Classifications
Eye Irrit. 2 - Skin Irrit. 2 - STOT SE 3
target_organs
Respiratory system
Storage Class
11 - Combustible Solids
wgk_germany
WGK 3
flash_point_f
Not applicable
flash_point_c
Not applicable
ppe
dust mask type N95 (US), Eyeshields, Gloves
Certificates of Analysis (COA)
Search for Certificates of Analysis (COA) by entering the products Lot/Batch Number. Lot and Batch Numbers can be found on a product’s label following the words ‘Lot’ or ‘Batch’.
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Articles
DNA damage and repair mechanism is vital for maintaining DNA integrity. Damage to cellular DNA is involved in mutagenesis, the development of cancer among others.
Cancer research has revealed that the classical model of carcinogenesis, a three step process consisting of initiation, promotion, and progression, is not complete.
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service