Efficient Gene Silencing using MISSION® TRC shRNA Lentiviral Vectors
Edward J. Weinstein, Betsy Boedeker, Roger Chiu, Andrea Spencer, Jenny Reade, Dave Cutter, Henry George
MilliporeSigma, St. Louis, MO, US
View pdf version (434KB)
Overview
The RNAi Consortium (TRC) is a collaborative effort among several academic laboratories (including the Broad Institute, the Whitehead Institute for Biomedical Research, the Dana Farber Cancer Institute, Massachusetts General Hospital, Massachusetts Institute of Technology, Harvard Medical School, Columbia University, and Washington University School of Medicine), as well as major biotechnology and pharmaceutical institutes, such as Eli Lilly, Novartis, and Bristol-Myers Squibb. The goal of the consortium is to validate methods and tools that will enable the scientific community to use RNAi in both mouse and human genes. To that end, reagents have been generated consisting of short hairpin sequences cloned into a lentiviral vector that can be used for a wide range of in vitro and in vivo studies. When completed, the Broad RNAi lentiviral library will consist of 150,000 custom-designed shRNAs, targeting 15,000 human (MISSION® TRC - Hs 1.0) and 15,000 mouse (MISSION® TRC - Mm 1.0) genes. Applications for this library will span the range from investigations into a single gene or pathway to genome-wide functional screens.

Background
While siRNAs have become the most common form of RNA inhibitors, they possess several qualities that may be less than optimal for a given experiment. The use of siRNAs requires a cell line that can be readily transfected. This trait is often missing from a number of cell types critical to today’s biomedical research, including primary cell cultures, stem cell lines, and even a wide array of human tumor cell lines. Another consideration is the transient nature of siRNA transfection. As cells that are successfully transfected with siRNA continue to divide, the concentration of siRNA is rapidly diluted, resulting in the gradual reaccumulation of target RNA. The long term silencing of a majority of cell lines can be achieved; however, through the use of shRNAs in a lentiviral transduction fashion.
We have examined the variable knockdown resulting after infection of the human lung tumor cell line A549 by four clones that are designed to target JAK1. Regulation of shRNA in A549 epithelial cells has been linked to regulation of JAK1 (Yokota et al., 2004) making manipulation of this gene particularly interesting in this cell line background. In addition to JAK1, we sought to examine the specificity of inhibition by shRNA using the gene AKT as an example. While all three isoforms of AKT (AKT1, AKT2, and AKT3) are expressed in A549 cells, inhibition of each individual isoform is believed to result in a different phenotype (Sithanandam et al., 2005). We transduced A549 cells with shRNAs to two of the three AKT isoforms, and examined down-regulation of the targeted isoform, as well as the two related, but non-targeted isoforms.
Results
The shRNAs targeting the JAK1 gene, when transduced into A549 cells, demonstrated varying levels of knockdown (Figure 1). Two of the constructs (TRCN0000003102 and TRCN000003105) reduced the endogenous level of JAK1 by more than 80%, while one construct (TRCN0000003104) inhibited about 75% of the message, and the fourth (TRCN0000003103) inhibited approximately 40%. With multiple shRNAs knocking down expression of JAK1 by 75% or more, there is a useful redundancy in reagents that can be utilized for proof of concept experiments. At the same time, there are shRNAs in the library that only partially reduce RNA levels of JAK1, allowing for examination of any hypomorphic phenotype associated with that gene. We utilized both GAPDH and cyclophilin as endogenous controls, and found the results to be very similar, indicating that either housekeeping gene is suitable for these experiments. The shRNAs found in the TRC library that target the various AKT isoforms yielded similar results in that infection of A549 with some shRNAs resulted in almost complete down-regulation of the target gene, while some yielded intermediary results (data not shown). Of particular note, however, is the specificity found within this library. The AKT2 targeting shRNA (TRCN0000000564) knocks out 75% of AKT2, but does not decrease the levels of AKT1 or AKT3 (Figure 2). Similarly, our representative shRNA targeting AKT3 (TRCN0000010187) reduces endogenous levels of that gene by about 60%, but does not significantly affect the levels of AKT1 or AKT2. Note that the levels of each endogenous gene are normalized to the levels seen after transduction of the empty control vector. The specificity of these reagents clearly allow the examination of the effects of reduction of each AKT isoform, which is of significant importance in light of the relevance of this gene to multiple cellular processes.
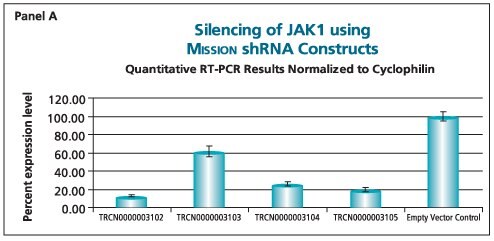
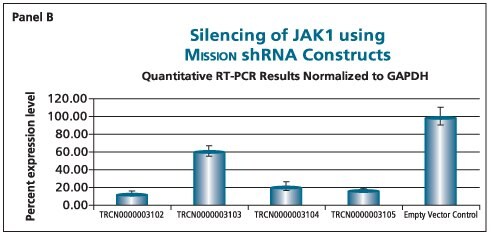
Figure 1.The Human Janus Kinase 1 (JAK1, NM_002227) MISSION® shRNA set (4 individual hairpins) was used to generate lentiviral particles in packaging cells. A549 cells were transduced with the lentiviral particles containing the shRNA sequences. 114 hours post-infection, total RNA was purified and analyzed with the appropriate TaqMan® Gene Expression assay. Results were normalized using GAPDH (Panel B) and cyclophilin (Panel A) quantitative RT-PCR results. The difference in Ct between infected samples and infected empty control cells, along with the efficiency of PCR, were used to generate percentage expression levels.
| shRNA Hairpin Sequence of Individual Clones | ||
|---|---|---|
| AKT1 | TRCN0000010174 | GGACTACCTGCACTCGGAGAA |
| AKT2 | TRCN0000000564 | CTTCGACTATCTCAAACTCCT |
| AKT3 | TRCN0000010187 | CTGCCTTGGACTATCTACATT |
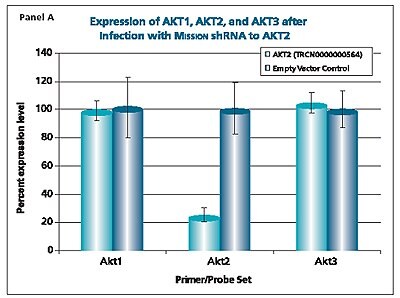
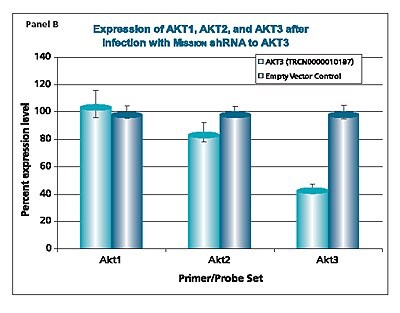
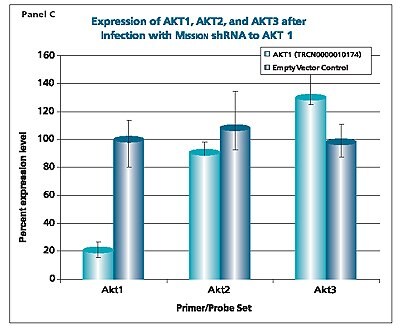
Figure 2.The Human v-akt murine thymoma viral oncogene homologs 1,2, and 3 (AKT1, NM_005163; AKT2, NM_001626; AKT3, NM_005465) MISSION® shRNA individual clones were used to generate lentiviral particles in packaging cells. A549 cells were transduced with the lentiviral particles containing the shRNA sequences. 114 hours post-infection, total RNA was purified and analyzed with the appropriate TaqMan® Gene Expression assay. The percent expression levels of AKT1, AKT2, and AKT3 were normalized against the empty vector control. Knockdown of AKT2 did not affect expression levels of AKT1 and AKT3 (Panel A). Knockdown of AKT3 did not affect expression levels of AKT1 and AKT2 (Panel B). Similarly, knockdown of AKT1 did not affect expression levels of AKT2 and AKT3 (Panel C).
Conclusion
RNA interference technology has been one of the key biological breakthroughs of the last decade and has revolutionized basic biology and gene function studies, but it also holds promise to revolutionize drug discovery and therapeutics. The MISSION® TRC shRNA libraries overcome the limitations of synthetic siRNAs by providing a system for long-term silencing and phenotypic observation, and the ability to generate lentiviral particles for infection and integration of primary cells, dividing cells, and nondividing cells. The libraries provide excellent specificity to target genes and high levels of transcript knockdown, making them a comprehensive set of tools for RNAi.
References
Contact Us
For questions about the library, pricing and quotes or other concerns, please email us.
MISSION is a trademark of Merck KGaA, Darmstadt, Germany and/or its affiliates. All other trademarks are the property of their respective owners. Detailed information on trademarks is available via publicly accessible resources. Label License.
To continue reading please sign in or create an account.
Don't Have An Account?