A6691
Amidase from Pseudomonas aeruginosa
recombinant, expressed in E. coli, buffered aqueous glycerol solution, hydroxamate transferase ≥200 units/mg protein (biuret)
Synonym(s):
Acrylamide Amidohydrolase, Acylase
Sign Into View Organizational & Contract Pricing
All Photos(1)
About This Item
CAS Number:
MDL number:
UNSPSC Code:
12352204
NACRES:
NA.54
Recommended Products
recombinant
expressed in E. coli
Quality Level
form
buffered aqueous glycerol solution
hydroxamate transferase activity
≥200 units/mg protein (biuret)
concentration
14 mg/mL
UniProt accession no.
storage temp.
−20°C
Gene Information
Pseudomonas aeruginosa PAO1 ... PA4163(880181)
Looking for similar products? Visit Product Comparison Guide
Application
Amidase from Pseudomonas aeruginosa has been used for testing its capability to hydrolyze ochratoxin A.
The importance of these hydrolases in biotechnology is growing rapidly, because their potential applications span through chemical and pharmaceutical industries as well as in bioremediation. Immobilized amidase can be used efficiently for production of acrylic acid from acrylamide, thus converting a toxic ambient contaminant into widely used industrial raw material. Amidases are potential treatments for human immunodeficiency virus and malaria. They may be used to eliminate metal ions in wastewater .
Biochem/physiol Actions
The amidase from Pseudomonas aeruginosa isa 6 × 38-kDa enzyme that catalyzes the hydrolysis of a small range of short aliphatic amides. Each amidase monomer is formed by a globular four-layer αββα sandwich domain with an additional 81-residue long C-terminal segment .
Unit Definition
One unit will convert 1.0 μmole of acetamide and hydroxylamine to acetohydroxamate and ammonia per min at pH 7.2 at 37 °C.
Physical form
Solution in 50% glycerol containing 7 mM 2-mercaptoethanol and phosphate buffer salt
signalword
Danger
hcodes
pcodes
Hazard Classifications
Resp. Sens. 1
Storage Class
12 - Non Combustible Liquids
wgk_germany
WGK 1
flash_point_f
Not applicable
flash_point_c
Not applicable
ppe
Eyeshields, Gloves, multi-purpose combination respirator cartridge (US)
Choose from one of the most recent versions:
Already Own This Product?
Find documentation for the products that you have recently purchased in the Document Library.
Customers Also Viewed
Substrate interaction with recombinant amidase from Pseudomonas aeruginosa during biocatalysis
Pacheco R, Karmali A
Biocatalysis and Biotransformation, 27(5/6), 367-376 (2009)
Jorge Andrade et al.
The Journal of biological chemistry, 282(27), 19598-19605 (2007-04-20)
Microbial amidases belong to the thiol nitrilases family and have potential biotechnological applications in chemical and pharmaceutical industries as well as in bioremediation. The amidase from Pseudomonas aeruginosa isa6 x 38-kDa enzyme that catalyzes the hydrolysis of a small range
Butyramide-utilizing mutants of Pseudomonas aeruginosa 8602 which produce an amidase with altered substrate specificity.
J E Brown et al.
Journal of general microbiology, 57(2), 273-285 (1969-08-01)
Arthur J L Cooper et al.
Comparative biochemistry and physiology. Part B, Biochemistry & molecular biology, 163(1), 108-120 (2012-05-23)
Many cancer cells have a strong requirement for glutamine. As an aid for understanding this phenomenon the (18)F-labeled 2S,4R stereoisomer of 4-fluoroglutamine [(2S,4R)4-FGln] was previously developed for in vivo positron emission tomography (PET). In the present work, comparative enzymological studies
Raffaella Gandolfi et al.
Bioorganic & medicinal chemistry letters, 22(16), 5283-5287 (2012-07-17)
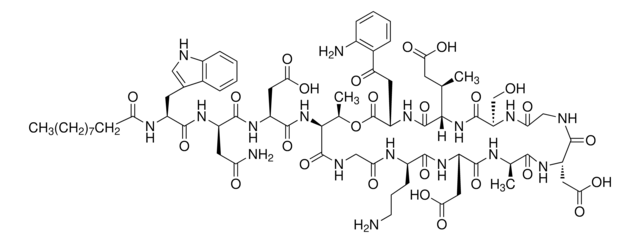
The chemoenzymatic deacylation of ramoplanin A2 is described for the first time: ramoplanin A2 was Boc-protected and hydrogenated to Boc-protected tetrahydroramoplanin, which was subsequently deacylated using an acylase from Actinoplanes utahensis NRRL 12052. The chemoenzymatic process proceeded with 80% overall
Our team of scientists has experience in all areas of research including Life Science, Material Science, Chemical Synthesis, Chromatography, Analytical and many others.
Contact Technical Service